Hands on with ChEMBL API
Posted on
*
•
10 minutes
•
2119 words
ChEMBL is a large-scale bioactivity database that collects information on the interactions between small molecules (such as drugs, compounds, or substances) and their biological targets. It stands for “Chemical Biology Database” and is maintained by the European Molecular Biology Laboratory (EMBL).
Researchers can use ChEMBL to search for specific compounds, investigate their interactions with biological targets (such as enzymes, receptors, or ion channels), and analyze bioactivity data to identify potential drug candidates or understand the mechanisms of action for existing drugs. Additionally, ChEMBL provides tools and APIs (Application Programming Interfaces) for accessing and querying the database programmatically, enabling integration with other bioinformatics and cheminformatics workflows.
import math
from pathlib import Path
from zipfile import ZipFile
from tempfile import TemporaryDirectory
import numpy as np
import pandas as pd
from rdkit.Chem import PandasTools
!pip install chembl- webresource- client
from chembl_webresource_client.new_client import new_client
from tqdm.auto import tqdm
DATA = Path. cwd() / 'data'
DATA. mkdir()
/Users/sasha/CADD/001_get_bioactivity_data_CHEMBL/data
resource objects for API access.
targets_api = new_client. target
compounds_api = new_client. molecule
bioactivities_api = new_client. activity
Get target data (EGFR kinase: UniProtID : P00533)¶
uniprot_id = "P00533"
# Get target information from ChEMBL for specified values only
targets = targets_api. get(target_components__accession= uniprot_id). only(
"target_chembl_id" , "organism" , "pref_name" , "target_type"
)
print (f 'The type of the targets is " { type (targets)} "' )
The type of the targets is "<class 'chembl_webresource_client.query_set.QuerySet'>"
targets = pd. DataFrame. from_records(targets)
targets
organism
pref_name
target_chembl_id
target_type
0
Homo sapiens
Epidermal growth factor receptor erbB1
CHEMBL203
SINGLE PROTEIN
1
Homo sapiens
Epidermal growth factor receptor erbB1
CHEMBL203
SINGLE PROTEIN
2
Homo sapiens
Epidermal growth factor receptor and ErbB2 (HE...
CHEMBL2111431
PROTEIN FAMILY
3
Homo sapiens
Epidermal growth factor receptor
CHEMBL2363049
PROTEIN FAMILY
4
Homo sapiens
MER intracellular domain/EGFR extracellular do...
CHEMBL3137284
CHIMERIC PROTEIN
5
Homo sapiens
Protein cereblon/Epidermal growth factor receptor
CHEMBL4523680
PROTEIN-PROTEIN INTERACTION
6
Homo sapiens
EGFR/PPP1CA
CHEMBL4523747
PROTEIN-PROTEIN INTERACTION
7
Homo sapiens
VHL/EGFR
CHEMBL4523998
PROTEIN-PROTEIN INTERACTION
8
Homo sapiens
Baculoviral IAP repeat-containing protein 2/Ep...
CHEMBL4802031
PROTEIN-PROTEIN INTERACTION
Restrict to first entry as our target of iterest
chembl_id = targets. iloc[0 ]. target_chembl_id
print (f " { chembl_id} " )
CHEMBL203
Fetch bioactivty data for the target_chembl_id : CHEMBL_203
# fetch the bioactivity data and filter it to only human proteins, IC50, exact measurement, binding data
bioactivities = bioactivities_api. filter(target_chembl_id= chembl_id, type = "IC50" , relation= "=" , assay_type= "B" ). only(
"activity_id" ,
"assay_chembl_id" ,
"assay_description" ,
"assay_type" ,
"molecule_chembl_id" ,
"type" ,
"standard_units" ,
"relation" ,
"standard_value" ,
"target_chembl_id" ,
"target_organism" ,
)
print (f "Length and type of bioactivities object: { len (bioactivities)} , { type (bioactivities)} " )
Length and type of bioactivities object: 10420, <class 'chembl_webresource_client.query_set.QuerySet'>
# Whats in here, look at first entry
bioactivities[0 ]
{'activity_id': 32260,
'assay_chembl_id': 'CHEMBL674637',
'assay_description': 'Inhibitory activity towards tyrosine phosphorylation for the epidermal growth factor-receptor kinase',
'assay_type': 'B',
'molecule_chembl_id': 'CHEMBL68920',
'relation': '=',
'standard_units': 'nM',
'standard_value': '41.0',
'target_chembl_id': 'CHEMBL203',
'target_organism': 'Homo sapiens',
'type': 'IC50',
'units': 'uM',
'value': '0.041'}
# Download into a data frame
bioactivities_df = pd. DataFrame. from_dict(bioactivities)
print (f "DataFrame shape: { bioactivities_df. shape} " )
bioactivities_df. head()
DataFrame shape: (10420, 13)
activity_id
assay_chembl_id
assay_description
assay_type
molecule_chembl_id
relation
standard_units
standard_value
target_chembl_id
target_organism
type
units
value
0
32260
CHEMBL674637
Inhibitory activity towards tyrosine phosphory...
B
CHEMBL68920
=
nM
41.0
CHEMBL203
Homo sapiens
IC50
uM
0.041
1
32267
CHEMBL674637
Inhibitory activity towards tyrosine phosphory...
B
CHEMBL69960
=
nM
170.0
CHEMBL203
Homo sapiens
IC50
uM
0.17
2
32680
CHEMBL677833
In vitro inhibition of Epidermal growth factor...
B
CHEMBL137635
=
nM
9300.0
CHEMBL203
Homo sapiens
IC50
uM
9.3
3
32770
CHEMBL674643
Inhibitory concentration of EGF dependent auto...
B
CHEMBL306988
=
nM
500000.0
CHEMBL203
Homo sapiens
IC50
uM
500.0
4
32772
CHEMBL674643
Inhibitory concentration of EGF dependent auto...
B
CHEMBL66879
=
nM
3000000.0
CHEMBL203
Homo sapiens
IC50
uM
3000.0
# units has different values
bioactivities_df["units" ]. unique()
array(['uM', 'nM', 'pM', 'M', "10'3 uM", "10'1 ug/ml", 'ug ml-1',
"10'-1microM", "10'1 uM", "10'-1 ug/ml", "10'-2 ug/ml", "10'2 uM",
"10'-3 ug/ml", "10'-2microM", '/uM', "10'-6g/ml", 'mM', 'umol/L',
'nmol/L', "10'-10M", "10'-7M", 'nmol', '10^-8M', 'µM'],
dtype=object)
# drop
bioactivities_df. drop(["units" , "value" ], axis= 1 , inplace= True )
bioactivities_df. head()
activity_id
assay_chembl_id
assay_description
assay_type
molecule_chembl_id
relation
standard_units
standard_value
target_chembl_id
target_organism
type
0
32260
CHEMBL674637
Inhibitory activity towards tyrosine phosphory...
B
CHEMBL68920
=
nM
41.0
CHEMBL203
Homo sapiens
IC50
1
32267
CHEMBL674637
Inhibitory activity towards tyrosine phosphory...
B
CHEMBL69960
=
nM
170.0
CHEMBL203
Homo sapiens
IC50
2
32680
CHEMBL677833
In vitro inhibition of Epidermal growth factor...
B
CHEMBL137635
=
nM
9300.0
CHEMBL203
Homo sapiens
IC50
3
32770
CHEMBL674643
Inhibitory concentration of EGF dependent auto...
B
CHEMBL306988
=
nM
500000.0
CHEMBL203
Homo sapiens
IC50
4
32772
CHEMBL674643
Inhibitory concentration of EGF dependent auto...
B
CHEMBL66879
=
nM
3000000.0
CHEMBL203
Homo sapiens
IC50
activity_id int64
assay_chembl_id object
assay_description object
assay_type object
molecule_chembl_id object
relation object
standard_units object
standard_value object
target_chembl_id object
target_organism object
type object
dtype: object
bioactivities_df = bioactivities_df. astype({"standard_value" : "float64" })
Data cleaning
Delete missing entries
Keep only entries with “standard_unit == nM”
Delete duplicates in molecule_chembl_id
Rename Columns
# Delete Missing values
bioactivities_df. dropna(axis= 0 , how= "any" , inplace= True )
print (f "DataFrame shape: { bioactivities_df. shape} " )
DataFrame shape: (10419, 11)
bioactivities_df = bioactivities_df[bioactivities_df["standard_units" ] == "nM" ]
bioactivities_df. drop_duplicates("molecule_chembl_id" , keep= "first" , inplace= True )
print (f "DataFrame shape: { bioactivities_df. shape} " )
DataFrame shape: (6823, 11)
bioactivities_df. reset_index(drop= True , inplace= True )
bioactivities_df. rename(columns= {"standard_value" : "IC50" , "standard_units" : "units" }, inplace= True )
bioactivities_df. head()
activity_id
assay_chembl_id
assay_description
assay_type
molecule_chembl_id
relation
units
IC50
target_chembl_id
target_organism
type
0
32260
CHEMBL674637
Inhibitory activity towards tyrosine phosphory...
B
CHEMBL68920
=
nM
41.0
CHEMBL203
Homo sapiens
IC50
1
32267
CHEMBL674637
Inhibitory activity towards tyrosine phosphory...
B
CHEMBL69960
=
nM
170.0
CHEMBL203
Homo sapiens
IC50
2
32680
CHEMBL677833
In vitro inhibition of Epidermal growth factor...
B
CHEMBL137635
=
nM
9300.0
CHEMBL203
Homo sapiens
IC50
3
32770
CHEMBL674643
Inhibitory concentration of EGF dependent auto...
B
CHEMBL306988
=
nM
500000.0
CHEMBL203
Homo sapiens
IC50
4
32772
CHEMBL674643
Inhibitory concentration of EGF dependent auto...
B
CHEMBL66879
=
nM
3000000.0
CHEMBL203
Homo sapiens
IC50
Fetch compound data (molecule_chembl_id) from ChEMBL
molecule_chembl_id = list (bioactivities_df["molecule_chembl_id" ])
compounds_provider = compounds_api. filter(molecule_chembl_id__in = molecule_chembl_id). only("molecule_chembl_id" , "molecule_structures" )
compounds = list (tqdm(compounds_provider))
compounds_df = pd. DataFrame. from_records(compounds)
print (f "DataFrame shape: { compounds_df. shape} " )
100%|██████████| 6823/6823 [06:32<00:00, 17.37it/s]
DataFrame shape: (6823, 2)
molecule_chembl_id
molecule_structures
0
CHEMBL6246
{'canonical_smiles': 'O=c1oc2c(O)c(O)cc3c(=O)o...
1
CHEMBL10
{'canonical_smiles': 'C[S+]([O-])c1ccc(-c2nc(-...
2
CHEMBL6976
{'canonical_smiles': 'COc1cc2c(cc1OC)Nc1ncn(C)...
3
CHEMBL7002
{'canonical_smiles': 'CC1(COc2ccc(CC3SC(=O)NC3...
4
CHEMBL414013
{'canonical_smiles': 'COc1cc2c(cc1OC)Nc1ncnc(O...
Preprocess and filter compound data
Remove entries with missing entries
Delete duplicate molecules (by molecule_chembl_id)
Get molecules with canonical SMILES
compounds_df. dropna(axis= 0 , how= "any" , inplace= True )
compounds_df. drop_duplicates("molecule_chembl_id" , keep= "first" , inplace= True )
# Check molecule_structures column
compounds_df. iloc[0 ]. molecule_structures
{'canonical_smiles': 'O=c1oc2c(O)c(O)cc3c(=O)oc4c(O)c(O)cc1c4c23',
'molfile': '\n RDKit 2D\n\n 22 25 0 0 0 0 0 0 0 0999 V2000\n -0.4750 -0.2417 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0\n 0.5750 0.3583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0\n -0.4750 -1.4792 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0\n 0.5750 1.6000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0\n -1.5333 0.3583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0\n 1.6292 -0.2417 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0\n 0.5750 -2.0875 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0\n -0.4750 2.2083 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0\n -1.5333 1.6000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0\n 1.6292 -1.4792 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0\n 1.6292 2.2083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0\n -1.5333 -2.0875 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0\n -2.5833 -0.2417 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0\n 2.6792 0.3583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0\n -2.5833 -1.4500 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0\n 2.6792 1.5625 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0\n -2.5625 2.2208 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0\n 2.6625 -2.1042 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0\n 1.6292 3.4000 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0\n -1.5333 -3.2917 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0\n -3.6375 -2.0125 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0\n 3.7375 2.1208 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0\n 2 1 1 0\n 3 1 1 0\n 4 2 2 0\n 5 1 2 0\n 6 2 1 0\n 7 3 1 0\n 8 9 1 0\n 9 5 1 0\n 10 7 1 0\n 11 4 1 0\n 12 3 2 0\n 13 5 1 0\n 14 6 2 0\n 15 13 2 0\n 16 14 1 0\n 17 9 2 0\n 18 10 2 0\n 19 11 1 0\n 20 12 1 0\n 21 15 1 0\n 22 16 1 0\n 12 15 1 0\n 4 8 1 0\n 6 10 1 0\n 11 16 2 0\nM END\n> <chembl_id>\nCHEMBL6246\n\n> <chembl_pref_name>\nundefined',
'standard_inchi': 'InChI=1S/C14H6O8/c15-5-1-3-7-8-4(14(20)22-11(7)9(5)17)2-6(16)10(18)12(8)21-13(3)19/h1-2,15-18H',
'standard_inchi_key': 'AFSDNFLWKVMVRB-UHFFFAOYSA-N'}
compounds_df. iloc[0 ]. molecule_structures. keys()
dict_keys(['canonical_smiles', 'molfile', 'standard_inchi', 'standard_inchi_key'])
# Keep only canonical_smiles
canonical_smiles = []
for i, compounds in compounds_df. iterrows():
try :
canonical_smiles. append(compounds["molecule_structures" ]["canonical_smiles" ])
except KeyError:
canonical_smiles. append(None )
compounds_df["smiles" ] = canonical_smiles
compounds_df. shape
(6816, 3)
molecule_chembl_id
molecule_structures
smiles
0
CHEMBL6246
{'canonical_smiles': 'O=c1oc2c(O)c(O)cc3c(=O)o...
O=c1oc2c(O)c(O)cc3c(=O)oc4c(O)c(O)cc1c4c23
1
CHEMBL10
{'canonical_smiles': 'C[S+]([O-])c1ccc(-c2nc(-...
C[S+]([O-])c1ccc(-c2nc(-c3ccc(F)cc3)c(-c3ccncc...
2
CHEMBL6976
{'canonical_smiles': 'COc1cc2c(cc1OC)Nc1ncn(C)...
COc1cc2c(cc1OC)Nc1ncn(C)c(=O)c1C2
3
CHEMBL7002
{'canonical_smiles': 'CC1(COc2ccc(CC3SC(=O)NC3...
CC1(COc2ccc(CC3SC(=O)NC3=O)cc2)CCCCC1
4
CHEMBL414013
{'canonical_smiles': 'COc1cc2c(cc1OC)Nc1ncnc(O...
COc1cc2c(cc1OC)Nc1ncnc(O)c1C2
# Are there missing smiles?
compounds_df[compounds_df["smiles" ]. isnull()]
molecule_chembl_id
molecule_structures
smiles
compounds_df. drop("molecule_structures" , axis= 1 , inplace= True )
compounds_df. head()
molecule_chembl_id
smiles
0
CHEMBL6246
O=c1oc2c(O)c(O)cc3c(=O)oc4c(O)c(O)cc1c4c23
1
CHEMBL10
C[S+]([O-])c1ccc(-c2nc(-c3ccc(F)cc3)c(-c3ccncc...
2
CHEMBL6976
COc1cc2c(cc1OC)Nc1ncn(C)c(=O)c1C2
3
CHEMBL7002
CC1(COc2ccc(CC3SC(=O)NC3=O)cc2)CCCCC1
4
CHEMBL414013
COc1cc2c(cc1OC)Nc1ncnc(O)c1C2
Merge dataframes
Index(['activity_id', 'assay_chembl_id', 'assay_description', 'assay_type',
'molecule_chembl_id', 'relation', 'units', 'IC50', 'target_chembl_id',
'target_organism', 'type'],
dtype='object')
Index(['molecule_chembl_id', 'smiles'], dtype='object')
print (f "Bioactivities filtered: { bioactivities_df. shape[0 ]} " )
print (f "Compounds filtered: { compounds_df. shape[0 ]} " )
Bioactivities filtered: 6823
Compounds filtered: 6816
# Merge DataFrames
output_df = pd. merge(
bioactivities_df[["molecule_chembl_id" , "IC50" , "units" ]],
compounds_df,
on= "molecule_chembl_id" ,
)
# Reset row indices
output_df. reset_index(drop= True , inplace= True )
print (f "Dataset with { output_df. shape[0 ]} entries." )
Dataset with 6816 entries.
molecule_chembl_id object
IC50 float64
units object
smiles object
dtype: object
molecule_chembl_id
IC50
units
smiles
0
CHEMBL68920
41.0
nM
Cc1cc(C)c(/C=C2\C(=O)Nc3ncnc(Nc4ccc(F)c(Cl)c4)...
1
CHEMBL69960
170.0
nM
Cc1cc(C(=O)N2CCOCC2)[nH]c1/C=C1\C(=O)Nc2ncnc(N...
2
CHEMBL137635
9300.0
nM
CN(c1ccccc1)c1ncnc2ccc(N/N=N/Cc3ccccn3)cc12
3
CHEMBL306988
500000.0
nM
CC(=C(C#N)C#N)c1ccc(NC(=O)CCC(=O)O)cc1
4
CHEMBL66879
3000000.0
nM
O=C(O)/C=C/c1ccc(O)cc1
5
CHEMBL77085
96000.0
nM
N#CC(C#N)=Cc1cc(O)ccc1[N+](=O)[O-]
6
CHEMBL443268
5310.0
nM
Cc1cc(C(=O)NCCN2CCOCC2)[nH]c1/C=C1\C(=O)N(C)c2...
7
CHEMBL76979
264000.0
nM
COc1cc(/C=C(\C#N)C(=O)O)cc(OC)c1O
8
CHEMBL76589
125.0
nM
N#CC(C#N)=C(N)/C(C#N)=C/c1ccc(O)cc1
9
CHEMBL76904
35000.0
nM
N#CC(C#N)=Cc1ccc(O)c(O)c1
Convert IC50 to pIC50
def convert_ic50_to_pic50 (IC50_value):
pIC50_value = 9 - math. log10(IC50_value)
return pIC50_value
# Apply conversion to each row of the compounds DataFrame
output_df["pIC50" ] = output_df. apply(lambda x: convert_ic50_to_pic50(x. IC50), axis= 1 )
molecule_chembl_id
IC50
units
smiles
pIC50
0
CHEMBL68920
41.0
nM
Cc1cc(C)c(/C=C2\C(=O)Nc3ncnc(Nc4ccc(F)c(Cl)c4)...
7.387216
1
CHEMBL69960
170.0
nM
Cc1cc(C(=O)N2CCOCC2)[nH]c1/C=C1\C(=O)Nc2ncnc(N...
6.769551
2
CHEMBL137635
9300.0
nM
CN(c1ccccc1)c1ncnc2ccc(N/N=N/Cc3ccccn3)cc12
5.031517
3
CHEMBL306988
500000.0
nM
CC(=C(C#N)C#N)c1ccc(NC(=O)CCC(=O)O)cc1
3.301030
4
CHEMBL66879
3000000.0
nM
O=C(O)/C=C/c1ccc(O)cc1
2.522879
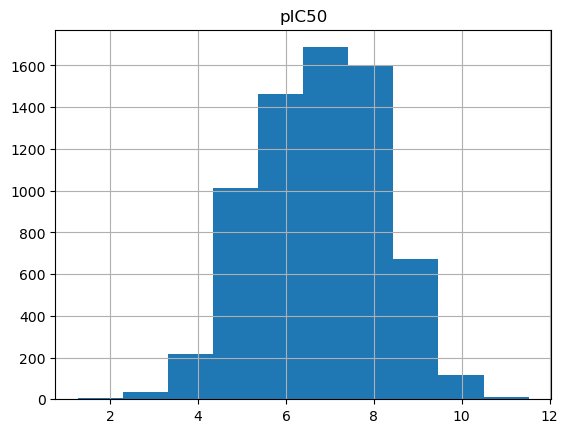
output_df. hist(column= "pIC50" )
array([[<Axes: title={'center': 'pIC50'}>]], dtype=object)
add a column for RDKit molecule objects
# Add molecule column
PandasTools. AddMoleculeColumnToFrame(output_df, smilesCol= "smiles" )
# Sort molecules by pIC50
output_df. sort_values(by= "pIC50" , ascending= False , inplace= True )
# Reset index
output_df. reset_index(drop= True , inplace= True )
#output_df.drop("smiles", axis=1).head(10)
molecule_chembl_id
IC50
units
smiles
pIC50
ROMol
0
CHEMBL63786
0.003
nM
Brc1cccc(Nc2ncnc3cc4ccccc4cc23)c1
11.522879
<rdkit.Chem.rdchem.Mol object at 0x1769d6810>
1
CHEMBL53711
0.006
nM
CN(C)c1cc2c(Nc3cccc(Br)c3)ncnc2cn1
11.221849
<rdkit.Chem.rdchem.Mol object at 0x1769cdfc0>
2
CHEMBL35820
0.006
nM
CCOc1cc2ncnc(Nc3cccc(Br)c3)c2cc1OCC
11.221849
<rdkit.Chem.rdchem.Mol object at 0x1769c1150>
3
CHEMBL53753
0.008
nM
CNc1cc2c(Nc3cccc(Br)c3)ncnc2cn1
11.096910
<rdkit.Chem.rdchem.Mol object at 0x1769c6dc0>
4
CHEMBL66031
0.008
nM
Brc1cccc(Nc2ncnc3cc4[nH]cnc4cc23)c1
11.096910
<rdkit.Chem.rdchem.Mol object at 0x1769c8350>
5
CHEMBL176582
0.010
nM
Cn1cnc2cc3ncnc(Nc4cccc(Br)c4)c3cc21
11.000000
<rdkit.Chem.rdchem.Mol object at 0x1769d66c0>
6
CHEMBL174426
0.025
nM
Cn1cnc2cc3c(Nc4cccc(Br)c4)ncnc3cc21
10.602060
<rdkit.Chem.rdchem.Mol object at 0x1769d6e30>
7
CHEMBL29197
0.025
nM
COc1cc2ncnc(Nc3cccc(Br)c3)c2cc1OC
10.602060
<rdkit.Chem.rdchem.Mol object at 0x1769c0dd0>
8
CHEMBL1243316
0.030
nM
C#CCNC/C=C/C(=O)Nc1cc2c(Nc3ccc(F)c(Cl)c3)c(C#N...
10.522879
<rdkit.Chem.rdchem.Mol object at 0x1769fa7a0>
9
CHEMBL363815
0.037
nM
C=CC(=O)Nc1ccc2ncnc(Nc3cc(Cl)c(Cl)cc3F)c2c1
10.431798
<rdkit.Chem.rdchem.Mol object at 0x1769dcb30>
output_df. to_csv(DATA / "EGFR_compounds.csv" )