GRU Regression for Molecular Property Prediction
Posted on * • 8 minutes • 1679 words
Gated Recurrent Units (GRU)
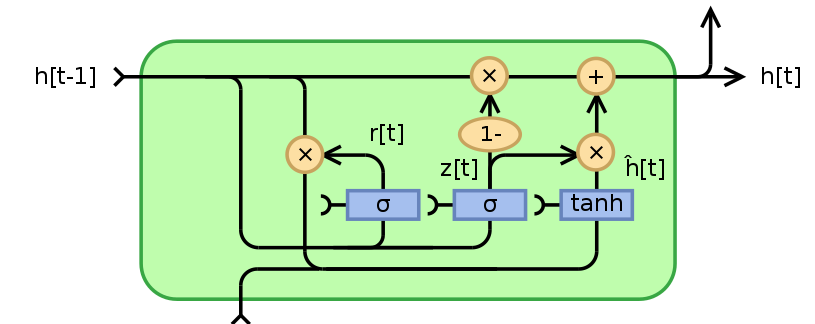
Gated Recurrent Unit (GRU) is a type of recurrent neural network (RNN) that was introduced by Cho et al. in 2014. It uses gating mechanisms to selectively update the hidden state of the network at each time step, allowing them to effectively model sequential data such as time series, natural language, and speech.
The network process sequential data by passing the hidden state from one time step to the next using gating mechanisms.
-
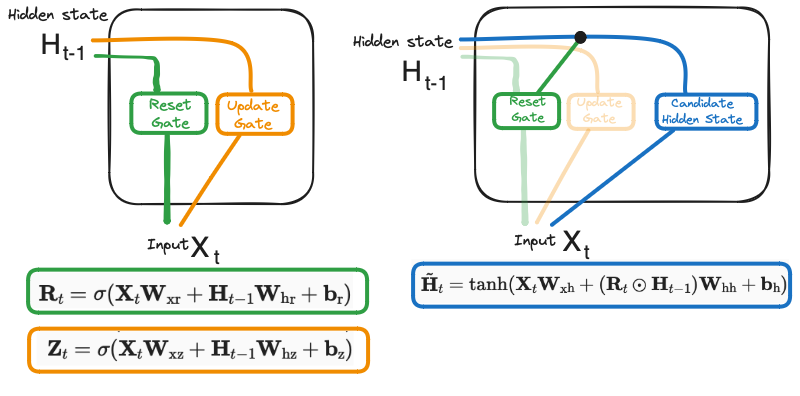
Reset Gate - identifies the unnecessary information and what information to delete at the specific timestamp.
-
Update Gate - identifies what current GRU cell will pass information to the next GRU cell thus, keeping track of the most important information.
-
Current Memory Gate or Candidate Hidden State Candidate Hidden State is used to determine the information stored from the past. This is generally called the memory component in a GRU cell.
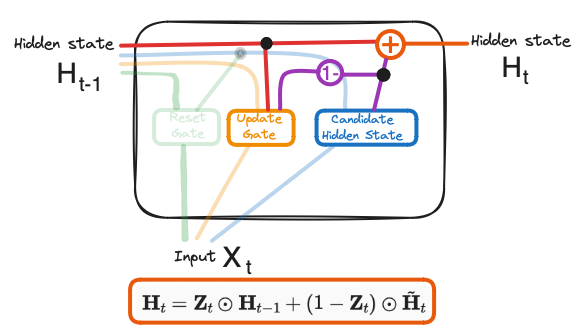
- New Hidden State - the new hidden state and depends on the update gate and candidate hidden state. whenever $\boldsymbol{Z}_t$ is $0$, the information at the previously hidden layer gets forgotten. It is updated with the value of the new candidate hidden layer. If $\boldsymbol{Z}_t$ is $1$, then the information from the previously hidden layer is maintained. This is how the most relevant information is passed from one state to the next.
Using GRU for Regression on qm9 dataset.
import os
import re
from collections import Counter
from pathlib import Path
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import torch
import torch.nn as nn
from torch.nn.utils.rnn import pad_sequence
from torch.utils.data import DataLoader, Dataset, Subset, TensorDataset
# Use cuda if available
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
# seed random generator
_ = torch.manual_seed(42)
Dataset
QM9_CSV_URL = "https://deepchemdata.s3-us-west-1.amazonaws.com/datasets/qm9.csv"
df = pd.read_csv(QM9_CSV_URL)
df.head()
| mol_id | smiles | A | B | C | mu | alpha | homo | lumo | gap | ... | zpve | u0 | u298 | h298 | g298 | cv | u0_atom | u298_atom | h298_atom | g298_atom | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | gdb_1 | C | 157.71180 | 157.709970 | 157.706990 | 0.0000 | 13.21 | -0.3877 | 0.1171 | 0.5048 | ... | 0.044749 | -40.478930 | -40.476062 | -40.475117 | -40.498597 | 6.469 | -395.999595 | -398.643290 | -401.014647 | -372.471772 |
| 1 | gdb_2 | N | 293.60975 | 293.541110 | 191.393970 | 1.6256 | 9.46 | -0.2570 | 0.0829 | 0.3399 | ... | 0.034358 | -56.525887 | -56.523026 | -56.522082 | -56.544961 | 6.316 | -276.861363 | -278.620271 | -280.399259 | -259.338802 |
| 2 | gdb_3 | O | 799.58812 | 437.903860 | 282.945450 | 1.8511 | 6.31 | -0.2928 | 0.0687 | 0.3615 | ... | 0.021375 | -76.404702 | -76.401867 | -76.400922 | -76.422349 | 6.002 | -213.087624 | -213.974294 | -215.159658 | -201.407171 |
| 3 | gdb_4 | C#C | 0.00000 | 35.610036 | 35.610036 | 0.0000 | 16.28 | -0.2845 | 0.0506 | 0.3351 | ... | 0.026841 | -77.308427 | -77.305527 | -77.304583 | -77.327429 | 8.574 | -385.501997 | -387.237686 | -389.016047 | -365.800724 |
| 4 | gdb_5 | C#N | 0.00000 | 44.593883 | 44.593883 | 2.8937 | 12.99 | -0.3604 | 0.0191 | 0.3796 | ... | 0.016601 | -93.411888 | -93.409370 | -93.408425 | -93.431246 | 6.278 | -301.820534 | -302.906752 | -304.091489 | -288.720028 |
5 rows × 21 columns
class SmilesTokenizer(object):
"""
A simple regex-based tokenizer adapted from the deepchem smiles_tokenizer package.
SMILES regex pattern for the tokenization is designed by Schwaller et. al., ACS Cent. Sci 5 (2019)
"""
def __init__(self):
self.regex_pattern = (
r"(\[[^\]]+]|Br?|Cl?|N|O|S|P|F|I|b|c|n|o|s|p|\(|\)|\."
r"|=|#|-|\+|\\|\/|:|~|@|\?|>>?|\*|\$|\%[0-9]{2}|[0-9])"
)
self.regex = re.compile(self.regex_pattern)
def tokenize(self, smiles):
"""
Tokenizes SMILES string.
"""
tokens = [token for token in self.regex.findall(smiles)]
return tokens
def build_vocab(smiles_list, tokenizer, max_vocab_size):
"""
Builds a vocabulary of N=max_vocab_size most common tokens from list of SMILES strings.
-------
Dict[str, int]
A dictionary that defines mapping of a token to its index in the vocabulary.
"""
tokenized_smiles = [tokenizer.tokenize(s) for s in smiles_list]
token_counter = Counter(c for s in tokenized_smiles for c in s)
tokens = [token for token, _ in token_counter.most_common(max_vocab_size)]
vocab = {token: idx for idx, token in enumerate(tokens)}
return vocab
def smiles_to_ohe(smiles, tokenizer, vocab):
"""
Transforms SMILES string to one-hot encoding representation.
Returns - Tensor
"""
unknown_token_id = len(vocab) - 1
token_ids = [vocab.get(token, unknown_token_id) for token in tokenizer.tokenize(smiles)]
ohe = torch.eye(len(vocab))[token_ids]
return ohe
# Test above functions
tokenizer = SmilesTokenizer()
smiles = "C=CS"
print("SMILES string:\n\t", smiles)
print("Tokens:\n\t", ", ".join(tokenizer.tokenize(smiles)))
vocab = build_vocab([smiles], tokenizer, 3)
print("Vocab:\n\t", vocab)
print("One-Hot-Enc:\n", np.array(smiles_to_ohe(smiles, tokenizer, vocab)).T)
SMILES string:
C=CS
Tokens:
C, =, C, S
Vocab:
{'C': 0, '=': 1, 'S': 2}
One-Hot-Enc:
[[1. 0. 1. 0.]
[0. 1. 0. 0.]
[0. 0. 0. 1.]]
PreProcess Data
sample_size = 50000
n_train = 40000
n_test = n_val = 5000
# get a sample
df = df.sample(n=sample_size, axis=0, random_state=42)
# select columns from the data frame
smiles = df["smiles"].tolist()
y = df["mu"].to_numpy()
# build a vocab using the training data
max_vocab_size = 30
vocab = build_vocab(smiles[:n_train], tokenizer, max_vocab_size)
vocab_size = len(vocab)
# transform smiles to one-hot encoded tensors and apply padding
X = pad_sequence(
sequences=[smiles_to_ohe(smi, tokenizer, vocab) for smi in smiles],
batch_first=True,
padding_value=0,
)
# normalize the target using the training data
train_mean = y[:n_train].mean()
train_std = y[:n_train].std()
y = (y - train_mean) / train_std
Build Dataset
# build dataset
data = TensorDataset(X, torch.Tensor(y))
# define loaders
ids_train = np.arange(n_train)
ids_val = np.arange(n_val) + n_train
ids_test = np.arange(n_test) + n_train + n_val
train_loader = DataLoader(
Subset(data, ids_train),
batch_size=64,
shuffle=True,
generator=torch.Generator().manual_seed(42),
)
val_loader = DataLoader(
Subset(data, ids_val), batch_size=64, shuffle=True, generator=torch.Generator().manual_seed(42)
)
test_loader = DataLoader(
Subset(data, ids_test),
batch_size=1,
shuffle=False,
generator=torch.Generator().manual_seed(42),
)
Build Model
class GRURegressionModel(nn.Module):
"""GRU network with one recurrent layer"""
def __init__(self, input_size, hidden_size=32, num_layers=1):
"""
GRU network
Parameters
----------
input_size : int
The number of expected features in the input vector
hidden_size : int
The number of features in the hidden state
"""
super(GRURegressionModel, self).__init__()
self.hidden_size = hidden_size
self.num_layers = num_layers
self.gru = nn.GRU(input_size, hidden_size, num_layers=1, batch_first=True)
self.fc = nn.Linear(hidden_size, 1)
self.dropout = nn.Dropout(p=0.2)
def forward(self, x):
h0 = torch.zeros(self.num_layers, x.size(0), self.hidden_size).to(device)
out, hn = self.gru(x, h0)
out = out[:, -1]
out = self.dropout(out)
out = self.fc(out)
return out
Training Class
class ModelTrainer(object):
"""A class that provides training and validation infrastructure for the model and keeps track of training and validation metrics."""
def __init__(self, model, lr, name=None, clip_gradients=False):
"""
Initialization.
Parameters
----------
model : nn.Module
a model
lr : float
learning rate for one training step
"""
self.model = model
self.lr = lr
self.criterion = torch.nn.MSELoss()
self.optimizer = torch.optim.Adam(self.model.parameters(), self.lr)
self.clip_gradients = clip_gradients
self.model.to(device)
self.train_loss = []
self.batch_loss = []
self.val_loss = []
def _train_epoch(self, loader):
self.model.train()
epoch_loss = 0
batch_losses = []
for i, (X_batch, y_batch) in enumerate(loader):
X_batch, y_batch = X_batch.to(device), y_batch.to(device)
self.optimizer.zero_grad()
y_pred = self.model(X_batch)
loss = self.criterion(y_pred, y_batch.unsqueeze(1))
loss.backward()
if self.clip_gradients:
torch.nn.utils.clip_grad_norm_(self.model.parameters(), max_norm=1, norm_type=2)
self.optimizer.step()
epoch_loss += loss.item()
batch_losses.append(loss.item())
return epoch_loss / len(loader), batch_losses
def _eval_epoch(self, loader):
self.model.eval()
val_loss = 0
predictions = []
targets = []
with torch.no_grad():
for X_batch, y_batch in loader:
X_batch, y_batch = X_batch.to(device), y_batch.to(device)
y_pred = self.model(X_batch)
loss = self.criterion(y_pred, y_batch.unsqueeze(1))
val_loss += loss.item()
predictions.append(y_pred.detach().numpy())
targets.append(y_batch.unsqueeze(1).detach().numpy())
predictions = np.concatenate(predictions).flatten()
targets = np.concatenate(targets).flatten()
return val_loss / len(loader), predictions, targets
def train(self, train_loader, val_loader, n_epochs, print_every=10):
"""
Train the model
Parameters
----------
train_loader :
a dataloader with training data
val_loader :
a dataloader with training data
n_epochs :
number of epochs to train for
"""
for e in range(n_epochs):
train_loss, train_loss_batches = self._train_epoch(train_loader)
val_loss, _, _ = self._eval_epoch(test_loader)
self.batch_loss += train_loss_batches
self.train_loss.append(train_loss)
self.val_loss.append(val_loss)
if e % print_every == 0:
print(f"Epoch {e+0:03} | train_loss: {train_loss:.5f} | val_loss: {val_loss:.5f}")
def validate(self, val_loader):
"""
Validate the model
Parameters
----------
val_loader :
a dataloader with training data
Returns
-------
Tuple[list, list, list]
Loss, y_predicted, y_target for each datapoint in val_loader.
"""
loss, y_pred, y_targ = self._eval_epoch(val_loader)
return loss, y_pred, y_targ
model_gru = ModelTrainer(
model=GRURegressionModel(vocab_size, hidden_size=32),
lr=1e-3,
)
model_gru.train(train_loader, val_loader, 51)
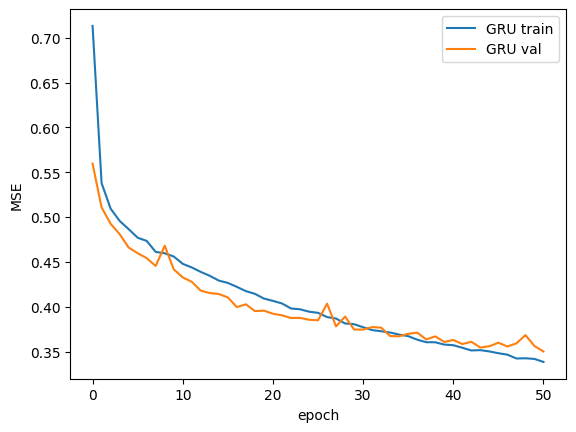
Epoch 000 | train_loss: 0.71333 | val_loss: 0.55967
Epoch 010 | train_loss: 0.44784 | val_loss: 0.43269
Epoch 020 | train_loss: 0.40651 | val_loss: 0.39215
Epoch 030 | train_loss: 0.37712 | val_loss: 0.37454
Epoch 040 | train_loss: 0.35705 | val_loss: 0.36291
Epoch 050 | train_loss: 0.33840 | val_loss: 0.35006
Loss Checking and Evaluation
_ = plt.plot(model_gru.train_loss, label=f"GRU train")
_ = plt.plot(model_gru.val_loss, label=f"GRU val")
_ = plt.xlabel("epoch")
_ = plt.ylabel("MSE")
_ = plt.legend()
Reference: Talktorial on Cheminformatics: T034 by Volkamer Lab