Morgan fingerprint and Tanimoto similarity
Posted on * • 2 minutes • 323 words
Morgan Fingerprint
Morgan fingerprints, also known as Morgan circular fingerprints or ECFP (Extended Connectivity Fingerprints), are a type of molecular fingerprint commonly used in cheminformatics and computational chemistry. They encode the molecular structure of a compound based on its connectivity to neighboring atoms within a defined radius. Morgan fingerprints are particularly useful in similarity searching, virtual screening, and quantitative structure-activity relationship (QSAR) studies.
Tanimoto similarity
Tanimoto similarity is often used to measure the similarity between two chemical compounds represented by their fingerprints. It calculates the similarity as the ratio of the intersection of the bits set to 1 in the fingerprints to the union of the bits set to 1. The Tanimoto coefficient ranges from 0 (no similarity) to 1 (complete similarity).
import urllib
import os
from rdkit import Chem
from rdkit.Chem import AllChem
from rdkit import DataStructs
from rdkit.Chem import Draw
import pandas as pd
import wget
# download smiles file
url = ("http://files.docking.org/2D/AA/AAAA.smi")
filename = wget.download(url)
100% [..............................................................................] 86110 / 86110
def process_smiles_file(filename):
smiles = {}
with open(filename, "r") as infile:
infile.readline() # skip header
for line in infile:
parts = line.split()
m = Chem.MolFromSmiles(parts[0])
if m is None:
continue
smiles[parts[1]] = m
return smiles
molecules = process_smiles_file("./AAAA.smi")
Build Fingerprint and run similarity
# Randomly use first molecule for the query fingerprint
fp_query = AllChem.GetMorganFingerprintAsBitVect(molecules[list(molecules.keys())[0]], 2)
# compute Tanimoto similarity for all molecules in our file
similarities = {}
for moleculeKey in molecules.keys():
fp2 = AllChem.GetMorganFingerprintAsBitVect(molecules[moleculeKey], 2)
similarity = DataStructs.FingerprintSimilarity(fp_query, fp2)
similarities[moleculeKey] = similarity
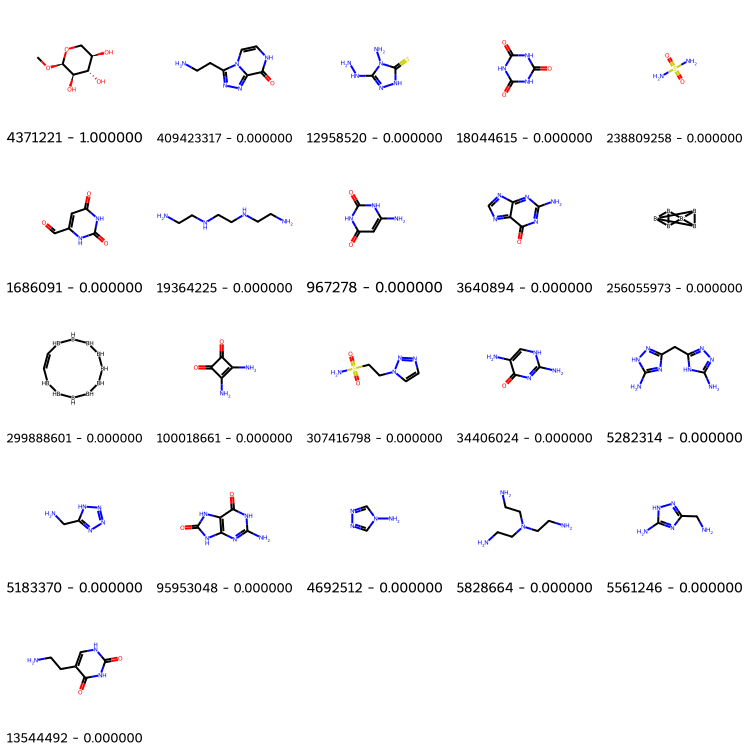
# get top 20 similar molecules
top20 = sorted(similarities, key=similarities.get, reverse=True)[:20]
top20.insert(0, list(molecules.keys())[0]) # this is the query molecule
# get bottom 20 similar molecules
bottom20 = sorted(similarities, key=similarities.get, reverse=False)[:20]
bottom20.insert(0, list(molecules.keys())[0]) # this is the query molecule
Draw.MolsToGridImage([molecules[x] for x in top20], molsPerRow=5, subImgSize=(150, 150),
legends=["%s - %f" % (x, similarities[x]) for x in top20])
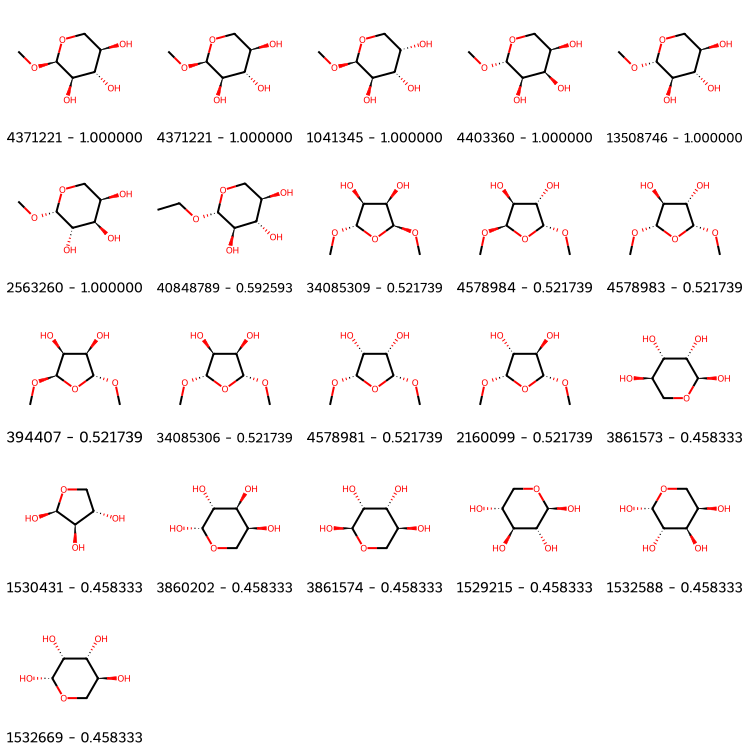
# draw top20 dissimilar molecules
Draw.MolsToGridImage([molecules[x] for x in bottom20], molsPerRow=5, subImgSize=(150, 150),
legends=["%s - %f" % (x, similarities[x]) for x in bottom20])