NetworkX - Visualizing PytorchGeometric Datasets
Posted on * • 9 minutes • 1888 words
import networkx as nx
In NetworkX, nodes can be any hashable object (eg., text, image, an XML object, another graph, a customized node object)
NetworkX includes many graph generator functions and facilities to read and write graphs in many formats.
- add_node
- add_nodes_from
- add_edge
- add_edges_from
- remove_node etc..
Create a graph
G = nx.Graph()
# nx.draw(G) #=> will be empty canvas
# add one node at a time,
G.add_node(1)
# nx.draw(G)
# Add from a collection
G.add_nodes_from([2,3,4,5])
#nx.draw(G)
G.add_edge(5,1)
#nx.draw(G)
G.add_nodes_from([6,7,8])
G.add_edges_from([(6,7),(7,8), (1,4), (2,3), (3,6),(5,6)])
nx.draw(G)
print(f"Number of nodes = {G.number_of_nodes()}")
print(f"Number of edges = {G.number_of_edges()}")
print(G.edges)
print(G.nodes)
Number of nodes = 8
Number of edges = 7
[(1, 5), (1, 4), (2, 3), (3, 6), (5, 6), (6, 7), (7, 8)]
[1, 2, 3, 4, 5, 6, 7, 8]
Export graph as JSON
from networkx.readwrite import json_graph
json_data = json_graph.node_link_data(G)
json_data
{'directed': False,
'multigraph': False,
'graph': {},
'nodes': [{'id': 1},
{'id': 2},
{'id': 3},
{'id': 4},
{'id': 5},
{'id': 6},
{'id': 7},
{'id': 8}],
'links': [{'source': 1, 'target': 5},
{'source': 1, 'target': 4},
{'source': 2, 'target': 3},
{'source': 3, 'target': 6},
{'source': 5, 'target': 6},
{'source': 6, 'target': 7},
{'source': 7, 'target': 8}]}
Read a json graph
json_data_to_graph = json_graph.node_link_graph(json_data)
json_data_to_graph
<networkx.classes.graph.Graph at 0x7e7f04523580>
Generate Graphml
for line in nx.generate_graphml(G):
print(line)
<graphml xmlns="http://graphml.graphdrawing.org/xmlns" xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance" xsi:schemaLocation="http://graphml.graphdrawing.org/xmlns http://graphml.graphdrawing.org/xmlns/1.0/graphml.xsd">
<graph edgedefault="undirected">
<node id="1" />
<node id="2" />
<node id="3" />
<node id="4" />
<node id="5" />
<node id="6" />
<node id="7" />
<node id="8" />
<edge source="1" target="5" />
<edge source="1" target="4" />
<edge source="2" target="3" />
<edge source="3" target="6" />
<edge source="5" target="6" />
<edge source="6" target="7" />
<edge source="7" target="8" />
</graph>
</graphml>
Remove Nodes or Clear graph
G.remove_nodes_from([1,3])
G.clear()
More of it
G.add_edges_from([(1,2),(1,3)])
print(G.edges())
print(G.nodes())
nx.draw(G)
[(1, 2), (1, 3)]
[1, 2, 3]
G.add_node("spam") # adds node "spam"
G.add_nodes_from("spam") # adds 4 nodes: 's', 'p', 'a', 'm'
print(G.edges())
print(G.nodes())
#nx.draw(G)
[(1, 2), (1, 3)]
[1, 2, 3, 'spam', 's', 'p', 'a', 'm']
Order, Density and Degree of Graph
Order => number of nodes Density for undirected graph:
$$ d = \frac{2e}{n(n-1)} $$ where $n$ is the number of nodes and $e$ is the number of edges
Degree : Returns a degree view
# Number of Nodes
G.order()
8
from networkx.classes.function import density
density(G)
0.07142857142857142
from networkx.classes.function import degree
degree(G, nbunch=None, weight=None)
DegreeView({1: 2, 2: 1, 3: 1, 'spam': 0, 's': 0, 'p': 0, 'a': 0, 'm': 0})
# In above graph G, degreeview shows 5 0's; 2 1's and 1 2;s
from networkx.classes.function import degree_histogram
degree_histogram(G)
[5, 2, 1]
from networkx.classes.function import neighbors
neighbors(G, 'spam')
<dict_keyiterator at 0x7e7f0442e070>
for node in G.nodes():
print(node, G.neighbors(node))
1 <dict_keyiterator object at 0x7e7f044dc400>
2 <dict_keyiterator object at 0x7e7f044dc400>
3 <dict_keyiterator object at 0x7e7f044dc400>
spam <dict_keyiterator object at 0x7e7f044dc400>
s <dict_keyiterator object at 0x7e7f044dc400>
p <dict_keyiterator object at 0x7e7f044dc400>
a <dict_keyiterator object at 0x7e7f044dc400>
m <dict_keyiterator object at 0x7e7f044dc400>
for node in G.nodes():
print(node, list(G.neighbors(node)))
1 [2, 3]
2 [1]
3 [1]
spam []
s []
p []
a []
m []
Graph Generators
https://networkx.org/documentation/stable/reference/generators.html
G.clear()
G = nx.binomial_tree(4)
nx.draw(G)
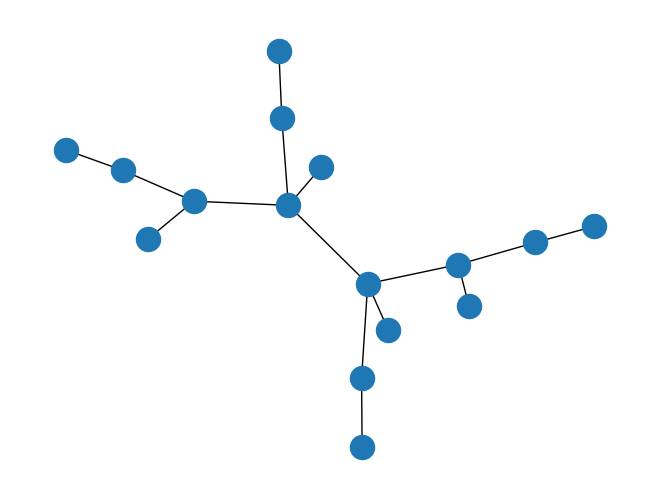
# default is 3x3 = 9 nodes
G = nx.sudoku_graph()
print(G.number_of_nodes())
print(G.number_of_edges())
print("--------------------")
A = nx.adjacency_matrix(G)
print(A.todense())
nx.draw(G)
81
810
--------------------
[[0 1 1 ... 0 0 0]
[1 0 1 ... 0 0 0]
[1 1 0 ... 0 0 0]
...
[0 0 0 ... 0 1 1]
[0 0 0 ... 1 0 1]
[0 0 0 ... 1 1 0]]
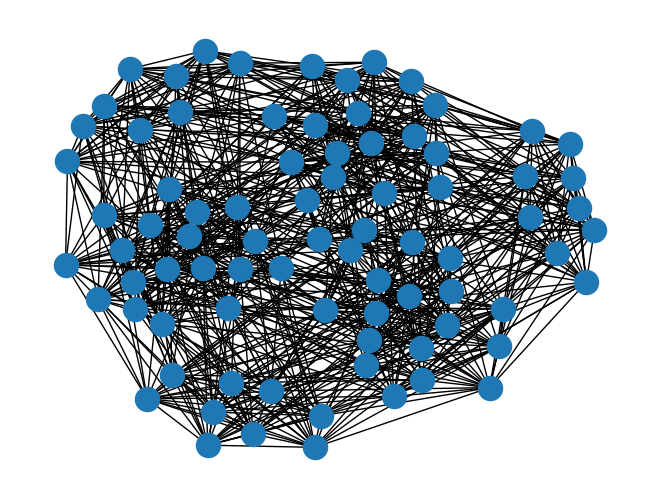
Directed Graphs
Neighbor And Adjacency
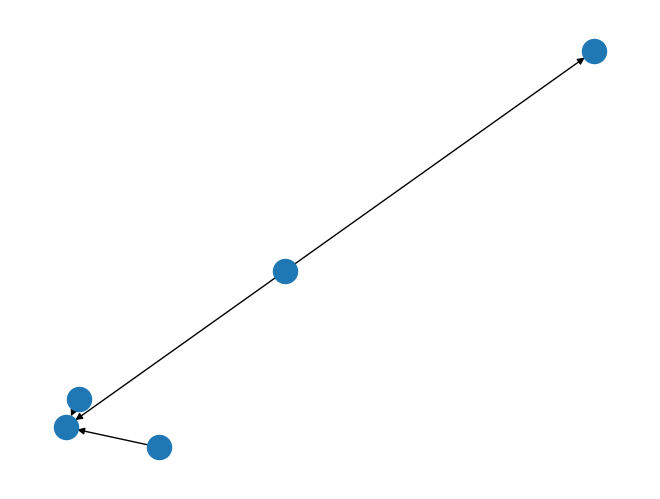
G = nx.DiGraph()
G.add_edge('a', 'b', weight=1)
G.add_edge('c', 'b', weight=5)
G.add_edge('m', 'n', weight=25)
G.add_edge('m', 'b', weight=50)
nx.draw(G)
print(nx.is_weighted(G))
print(nx.is_directed(G))
print(G.order())
print(G.number_of_edges())
print(G.number_of_nodes())
print(G.edges)
print(G.nodes)
True
True
5
4
5
[('a', 'b'), ('c', 'b'), ('m', 'n'), ('m', 'b')]
['a', 'b', 'c', 'm', 'n']
print([n for n in G.neighbors('a')])
print("===========")
for node in G.nodes():
print(node, list(G.neighbors(node)))
print("===========")
print([n for n in G.neighbors('m')])
['b']
===========
a ['b']
b []
c ['b']
m ['n', 'b']
n []
===========
['n', 'b']
for x in G.nodes:
print('Neighbors for ' + x + ':')
print([n for n in G.neighbors(x)])
Neighbors for a:
['b']
Neighbors for b:
[]
Neighbors for c:
['b']
Neighbors for m:
['n', 'b']
Neighbors for n:
[]
A = nx.adjacency_matrix(G)
print(A)
print("====")
print(A.todense())
(0, 1) 1
(2, 1) 5
(3, 1) 50
(3, 4) 25
====
[[ 0 1 0 0 0]
[ 0 0 0 0 0]
[ 0 5 0 0 0]
[ 0 50 0 0 25]
[ 0 0 0 0 0]]
# Is it self-looped
A.diagonal()
array([0, 0, 0, 0, 0])
# From G to numpy array gives A
A = nx.to_numpy_array(G)
print(A)
[[ 0. 1. 0. 0. 0.]
[ 0. 0. 0. 0. 0.]
[ 0. 5. 0. 0. 0.]
[ 0. 50. 0. 0. 25.]
[ 0. 0. 0. 0. 0.]]
Multiple Edge Attributes
import numpy as np
G = nx.Graph()
G.add_edge(0, 1, weight=10)
G.add_edge(1, 2, cost=5)
G.add_edge(2, 3, weight=3, cost=-4.0)
dtype = np.dtype([("weight", int), ("cost", float)])
# To create adjacency matrices from structured dtypes, use `weight=None`
A = nx.to_numpy_array(G, dtype=dtype, weight=None)
print("weight --------")
print(A["weight"])
print("cost --------")
print(A["cost"])
print(G.edges)
print(G.nodes)
weight --------
[[ 0 10 0 0]
[10 0 1 0]
[ 0 1 0 3]
[ 0 0 3 0]]
cost --------
[[ 0. 1. 0. 0.]
[ 1. 0. 5. 0.]
[ 0. 5. 0. -4.]
[ 0. 0. -4. 0.]]
[(0, 1), (1, 2), (2, 3)]
[0, 1, 2, 3]
G = nx.Graph(
[
("A", "B", {"cost": 1, "weight": 7}),
("C", "E", {"cost": 9, "weight": 10}),
]
)
print(G.edges)
print(G.nodes)
print("---------- #For entire graph ----------")
df = nx.to_pandas_edgelist(G)
print(df)
print("--------#for selected list BE------------")
df = nx.to_pandas_edgelist(G, nodelist=["B", "E"]) #for selected list
print(df)
print("--------------------")
df = nx.to_pandas_edgelist(G, nodelist=["A", "C"])
print(df)
print("--------------------")
df[["source", "target", "cost", "weight"]]
[('A', 'B'), ('C', 'E')]
['A', 'B', 'C', 'E']
---------- #For entire graph ----------
source target cost weight
0 A B 1 7
1 C E 9 10
--------#for selected list BE------------
source target cost weight
0 B A 1 7
1 E C 9 10
--------------------
source target cost weight
0 A B 1 7
1 C E 9 10
--------------------
# Only weights
A = nx.adjacency_matrix(G)
print(A.todense())
[[ 0 7 0 0]
[ 7 0 0 0]
[ 0 0 0 10]
[ 0 0 10 0]]
Viewing Datasets from Pytorch Geometry
1. Enzyme Dataset
!python -c "import torch; print(torch.__version__)"
2.1.0+cu121
!pip install torch-scatter -f https://data.pyg.org/whl/torch-2.1.0+cu121.html
!pip install torch-sparse -f https://data.pyg.org/whl/torch-2.1.0+cu121.html
!pip install torch-geometric
from torch_geometric.datasets import TUDataset
dataset = TUDataset(root='/tmp/ENZYMES', name='ENZYMES')
Downloading https://www.chrsmrrs.com/graphkerneldatasets/ENZYMES.zip
Processing...
Done!
print(f"{dataset} : {len(dataset)}")
print(f"Num classes : {dataset.num_classes}")
print(f"Num classes : {dataset.num_node_features}")
ENZYMES(600) : 600
Num classes : 6
Num classes : 3
data = dataset[0]
data
Data(edge_index=[2, 168], x=[37, 3], y=[1])
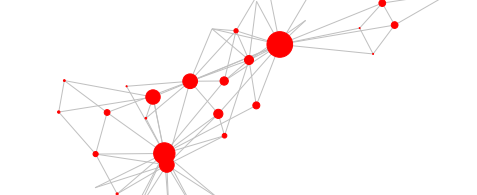
from torch_geometric.utils import to_networkx
print(type(data))
networkX_graph = to_networkx(data)
print(type(networkX_graph))
<class 'torch_geometric.data.data.Data'>
<class 'networkx.classes.digraph.DiGraph'>
import networkx as nx
nx.draw(networkX_graph)
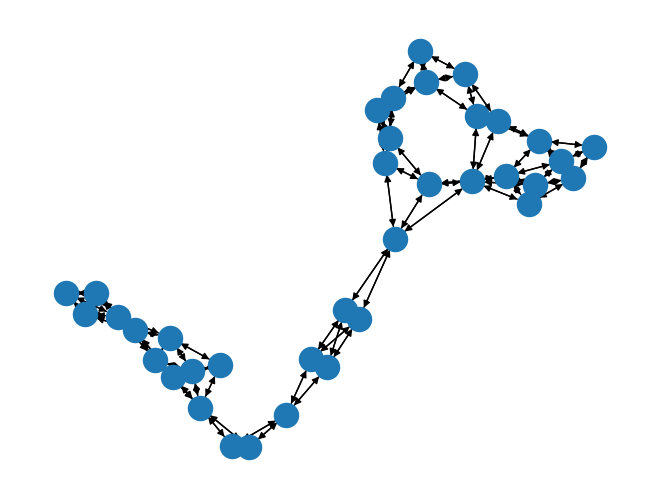
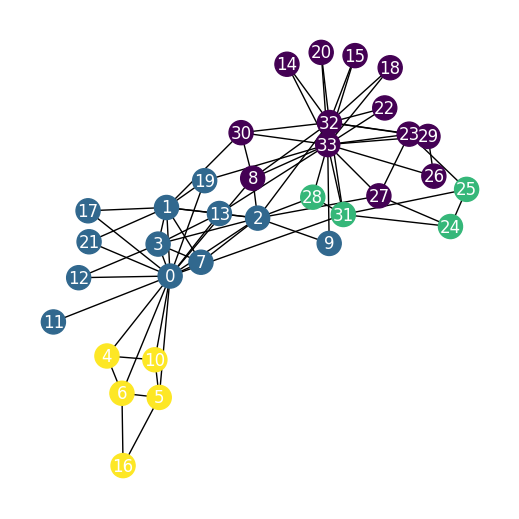
2. Karate Dataset
# Helper function for visualization.
%matplotlib inline
import matplotlib.pyplot as plt
from torch_geometric.datasets import KarateClub
dataset = KarateClub()
print(f'Dataset: {dataset}:')
print('======================')
print(f'Number of graphs: {len(dataset)}')
print(f'Number of features: {dataset.num_features}')
print(f'Number of classes: {dataset.num_classes}')
print(f'Number of Node Features: {dataset.num_node_features}')
print(f'Number of Edge Features: {dataset.num_edge_features}')
Dataset: KarateClub():
======================
Number of graphs: 1
Number of features: 34
Number of classes: 4
Number of Node Features: 34
Number of Edge Features: 0
- This dataset holds exactly one graph,
- Each node in this dataset is assigned a 34-dimensional feature vector (which uniquely describes the members of the karate club).
- The graph holds exactly 4 classes, which represent the community each node belongs to.
data = dataset[0] # Get the first graph object.
print(data)
# (1) The edge_index property holds the information about the graph connectivity, i.e., a tuple of source and destination node indices for each edge.
# (2) node features as x (each of the 34 nodes is assigned a 34-dim feature vector)
# (3) node labels as y (each node is assigned to exactly one class).
Data(x=[34, 34], edge_index=[2, 156], y=[34], train_mask=[34])
print('==============================================================')
# Gather some statistics about the graph.
print(f'Number of nodes: {data.num_nodes}')
print(f'Number of edges: {data.num_edges}')
print(f'Average node degree: {data.num_edges / data.num_nodes:.2f}')
print(f'Has isolated nodes: {data.has_isolated_nodes()}')
print(f'Has self-loops: {data.has_self_loops()}')
print(f'Is Directed: {data.is_directed()}')
print(f'Is undirected: {data.is_undirected()}')
print('==============================================================')
print(f'Edge weight: {data.edge_weight}')
print(f'Graph contains isolated nodes: {data.contains_isolated_nodes()}')
print('==============================================================')
print(f'Number of training nodes: {data.train_mask.sum()}')
print(f'Training node label rate: {int(data.train_mask.sum()) / data.num_nodes:.2f}')
==============================================================
Number of nodes: 34
Number of edges: 156
Average node degree: 4.59
Has isolated nodes: False
Has self-loops: False
Is Directed: False
Is undirected: True
==============================================================
Edge weight: None
Graph contains isolated nodes: False
==============================================================
Number of training nodes: 4
Training node label rate: 0.12
/usr/local/lib/python3.10/dist-packages/torch_geometric/deprecation.py:26: UserWarning: 'contains_isolated_nodes' is deprecated, use 'has_isolated_nodes' instead
warnings.warn(out)
data.to_dict()
{'x': tensor([[1., 0., 0., ..., 0., 0., 0.],
[0., 1., 0., ..., 0., 0., 0.],
[0., 0., 1., ..., 0., 0., 0.],
...,
[0., 0., 0., ..., 1., 0., 0.],
[0., 0., 0., ..., 0., 1., 0.],
[0., 0., 0., ..., 0., 0., 1.]]),
'edge_index': tensor([[ 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1,
1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 3,
3, 3, 3, 3, 3, 4, 4, 4, 5, 5, 5, 5, 6, 6, 6, 6, 7, 7,
7, 7, 8, 8, 8, 8, 8, 9, 9, 10, 10, 10, 11, 12, 12, 13, 13, 13,
13, 13, 14, 14, 15, 15, 16, 16, 17, 17, 18, 18, 19, 19, 19, 20, 20, 21,
21, 22, 22, 23, 23, 23, 23, 23, 24, 24, 24, 25, 25, 25, 26, 26, 27, 27,
27, 27, 28, 28, 28, 29, 29, 29, 29, 30, 30, 30, 30, 31, 31, 31, 31, 31,
31, 32, 32, 32, 32, 32, 32, 32, 32, 32, 32, 32, 32, 33, 33, 33, 33, 33,
33, 33, 33, 33, 33, 33, 33, 33, 33, 33, 33, 33],
[ 1, 2, 3, 4, 5, 6, 7, 8, 10, 11, 12, 13, 17, 19, 21, 31, 0, 2,
3, 7, 13, 17, 19, 21, 30, 0, 1, 3, 7, 8, 9, 13, 27, 28, 32, 0,
1, 2, 7, 12, 13, 0, 6, 10, 0, 6, 10, 16, 0, 4, 5, 16, 0, 1,
2, 3, 0, 2, 30, 32, 33, 2, 33, 0, 4, 5, 0, 0, 3, 0, 1, 2,
3, 33, 32, 33, 32, 33, 5, 6, 0, 1, 32, 33, 0, 1, 33, 32, 33, 0,
1, 32, 33, 25, 27, 29, 32, 33, 25, 27, 31, 23, 24, 31, 29, 33, 2, 23,
24, 33, 2, 31, 33, 23, 26, 32, 33, 1, 8, 32, 33, 0, 24, 25, 28, 32,
33, 2, 8, 14, 15, 18, 20, 22, 23, 29, 30, 31, 33, 8, 9, 13, 14, 15,
18, 19, 20, 22, 23, 26, 27, 28, 29, 30, 31, 32]]),
'y': tensor([1, 1, 1, 1, 3, 3, 3, 1, 0, 1, 3, 1, 1, 1, 0, 0, 3, 1, 0, 1, 0, 1, 0, 0,
2, 2, 0, 0, 2, 0, 0, 2, 0, 0]),
'train_mask': tensor([ True, False, False, False, True, False, False, False, True, False,
False, False, False, False, False, False, False, False, False, False,
False, False, False, False, True, False, False, False, False, False,
False, False, False, False])}
from IPython.display import Javascript # Restrict height of output cell.
display(Javascript('''google.colab.output.setIframeHeight(0, true, {maxHeight: 300})'''))
<IPython.core.display.Javascript object>
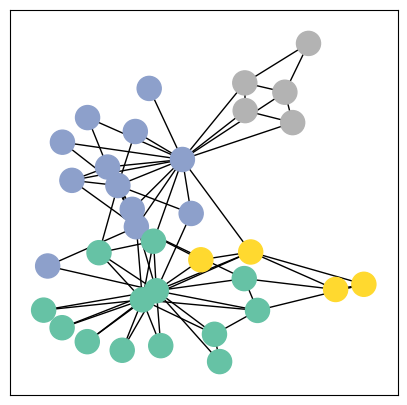
def visualize_graph(G, color):
plt.figure(figsize=(5,5))
plt.xticks([])
plt.yticks([])
nx.draw_networkx(G, pos=nx.spring_layout(G, seed=42), with_labels=False,
node_color=color, cmap="Set2")
plt.show()
karate_undirected_graph = to_networkx(data, to_undirected=True)
visualize_graph(karate_undirected_graph, color=data.y)
plt.figure(figsize=(5,5))
nx.draw(karate_undirected_graph, cmap=plt.get_cmap('viridis'), with_labels=True, node_color=data.y, font_color='white')
# 4 Classes are visible