Peptide Embedding for Unsupervised clustering
Posted on * • 10 minutes • 2076 words
How the peptides were encoded in an interesting paper titled: Unsupervised machine learning leads to an abiotic picomolar peptide ligand.
Different emebdeings that were used:
- one-hot encoding
- extended connectivity Fingerprints (ECFP_6) from RDKit represented each amino acid as a vector, where each index indicates the presence (1) or absence (0) of a specific molecular substructure
- physicochemical encoding based on 12 physicochemical properties.
- N-gramsencodingrepresentedtheentirepeptidebyitsungappedmotifs, irrespective of position.
- Latent embeddings of the entire peptide from the protein language model ESM-2.26. This is not included in this notebook.
Read data for target-specific binders, nonspecific and nonbinding peptides form affinity- selection mass-spectrometry (AS-MS)
import pandas as pd
# Inputs
file = './All Data Randomized C-L.csv'
df = pd.read_csv(file)
df
| Peptide | Label | ALC | |
|---|---|---|---|
| 0 | VWRDAEDYATFPK | Specific | NaN |
| 1 | TPDWDGPDYAAHK | Specific | NaN |
| 2 | EMDAPDYASWEPK | Specific | NaN |
| 3 | SVEDDAPDYADFK | Specific | NaN |
| 4 | MMDMDLQDYAGLK | Specific | NaN |
| ... | ... | ... | ... |
| 4099 | LLKYTDRHDYAWK | Specific | NaN |
| 4100 | DTDWPDYSSFLFK | Specific | NaN |
| 4101 | DSWEDYSSNTAAK | Unknown | NaN |
| 4102 | PQDLSEYAHSNNK | Nonspecific | NaN |
| 4103 | FLYDVHDYAFEHK | Specific | NaN |
4104 rows × 3 columns
# Isolates the X12 variable region from all peptides. This is specific to AS-MS
seq_list = [seq[0:12] for seq in df['Peptide']]
seq_list[-5:]
['LLKYTDRHDYAW',
'DTDWPDYSSFLF',
'DSWEDYSSNTAA',
'PQDLSEYAHSNN',
'FLYDVHDYAFEH']
import math
print(f'{len(seq_list)} peptides', flush=True)
print(f'If using UMAP, n_neighbors is recommended to be {math.floor(len(seq_list)*0.05)} with n_bors = {math.ceil(math.log2(len(seq_list)*0.05))}')
4104 peptides
If using UMAP, n_neighbors is recommended to be 205 with n_bors = 8
One-hot encoding
# Fingerprint encoding using RDKit
Std_All_AA_Smiles = {
'A' : 'CNC(=O)[C@H](C)NC(C)=O',
'C' : 'CNC(=O)[C@H](CS)NC(C)=O', # note that Cys is not used in AS-MS libraries for ths work, but is included for other use cases
'D' : 'CNC(=O)[C@H](CC(=O)O)NC(C)=O',
'E' : 'CNC(=O)[C@H](CCC(=O)O)NC(C)=O',
'F' : 'CNC(=O)[C@H](Cc1ccccc1)NC(C)=O',
'G' : 'CNC(=O)CNC(C)=O',
'H' : 'CNC(=O)[C@H](Cc1c[nH]cn1)NC(C)=O',
'I' : 'CC[C@H](C)[C@H](NC(C)=O)C(=O)NC', # note that Ile is not used in AS-MS libraries for ths work, but is included for other use cases
'K' : 'CNC(=O)[C@H](CCCCN)NC(C)=O',
'L' : 'CNC(=O)[C@H](CC(C)C)NC(C)=O',
'M' : 'CNC(=O)[C@H](CCSC)NC(C)=O',
'N' : 'CNC(=O)[C@H](CC(N)=O)NC(C)=O',
'P' : 'CNC(=O)[C@@H]1CCCN1C(C)=O',
'Q' : 'CNC(=O)[C@H](CCC(N)=O)NC(C)=O',
'R' : 'CNC(=O)[C@H](CCCNC(=N)N)NC(C)=O',
'S' : 'CNC(=O)[C@H](CO)NC(C)=O',
'T' : 'CNC(=O)[C@@H](NC(C)=O)[C@@H](C)O',
'V' : 'CNC(=O)[C@@H](NC(C)=O)C(C)C',
'W' : 'CNC(=O)[C@H](Cc1c[nH]c2ccccc12)NC(C)=O',
'Y' : 'CNC(=O)[C@H](Cc1ccc(O)cc1)NC(C)=O',
}
from sklearn import preprocessing
import numpy as np
# OneHot encoding function
enc_one = preprocessing.OneHotEncoder(handle_unknown='ignore')
enc_one.fit(np.array(list(Std_All_AA_Smiles.keys())).reshape(-1, 1))
def one_hot(sequence):
return enc_one.transform(np.array(list(sequence)).reshape(-1,1)).toarray().reshape(-1)
seq_list[0]
'VWRDAEDYATFP'
one_hot(seq_list[0])
array([0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0.,
1., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0.,
0., 0., 0., 0., 1., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0.,
0., 0., 0., 1., 0., 0., 0., 0., 0., 0., 0., 1., 0., 0., 0., 0., 0.,
0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 1., 0., 0., 0., 0.,
0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0.,
0., 1., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0.,
0., 0., 0., 1., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0.,
0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0.,
0., 0., 0., 0., 0., 0., 1., 1., 0., 0., 0., 0., 0., 0., 0., 0., 0.,
0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0.,
0., 0., 0., 0., 0., 0., 0., 0., 0., 1., 0., 0., 0., 0., 0., 0., 0.,
1., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0.,
0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 1., 0., 0., 0., 0., 0.,
0., 0.])
# DO it for all seqeunces
seq_rep = {}
seq_rep['OneHot'] = [one_hot(seq) for seq in seq_list]
# List to keep representation
seq_rep_list = ['OneHot', 'PhysProperties', 'Fingerprint', 'NGrams']
Fingerprint Encoding
from rdkit.Chem import AllChem as Chem
class Fingerprint_Generation:
def __init__(self, smiles,radius,nbits):
self.lookupfps = {}
for key, value in Std_All_AA_Smiles.items():
mol = Chem.MolFromSmiles(value)
fp = np.array(Chem.GetMorganFingerprintAsBitVect(mol,radius,nbits))
self.lookupfps[key] = fp
self.lookupfps[' '] = np.zeros(self.lookupfps['A'].shape)
self.lookupfps['x'] = np.ones(self.lookupfps['A'].shape)
def seq(self, seq):
fp = np.asarray([self.lookupfps[seq[i]] for i in range(len(seq))])
return fp
def fingerprint(sequence,NBITS,fp):
fp_seq = fp.seq(sequence)
return fp_seq.reshape(-1)
# Parameters
NBITS = 256 # For Fingerprint encoding, 256 was optimal
RADIUS = 3 # For Fingerprint encoding, ECFP_6
fp = Fingerprint_Generation(Std_All_AA_Smiles.keys(),RADIUS,NBITS)
seq_rep['Fingerprint'] = [fingerprint(seq,NBITS,fp) for seq in seq_list]
Using Physical Properties for Encoding
based on 12 physicochemical properties of its composite amino acids: hydrophilicity, flexibility, accessibility, turns scale, exposed surface, polarity, antigenic propensity, hydrophobicity, net charge index of the side chains, polarizability, solvent-accessible surface area, and side-chain volume.
For details See; Residue–Residue Interaction Prediction via Stacked Meta-Learning https://doi.org/10.3390/ijms22126393
# Extended physical properties database and encoding
extended_prop_aa = {
'A':[0.620140363440966,0.17686056841772,-0.188749006724763,-0.224734173518473,-0.0836274309924442,-0.553429077935438,-1.32955152740811,-1.38172110737113,-1.02627398746377,-0.509282137090254,-0.585795803359558,-0.781110425203712,-0.520029443227509,0.213921592667873],
'C':[0.290065653867549,0.066322713156645,-0.440414349024447,-0.226412219190666,-1.04998885579402,-0.476729477285148,-0.489295456136705,-0.77494270388318,-0.578373331393242,-0.427542694668279,-1.63430191054009,-1.21748496442366,0.881041618691234,2.14004615278222],
'D':[-0.900203753382047,1.42435922064985,1.57290838937303,-0.225916432969336,1.73759217728745,1.55580993994754,-0.724977037103076,-0.501892422313604,-0.698861227178295,0.712268419104827,0.73623363612894,0.746200462066116,1.58587491364422,-0.881976863948911],
'E':[-0.740167530558572,1.07695453268647,1.57290838937303,-0.224734173518473,1.47741794753318,2.03310948183212,-0.253613875170334,0.0940506525406323,-0.122614769075867,0.794007861526803,1.0097569684369,0.964387731676091,-0.623175779074287,-0.964999474298668],
'F':[1.1902694072496,-1.60753623793963,-1.1954103759235,-0.22355191406761,-1.16149209711728,-0.535290658862734,1.1707226358873,0.887196708528451,1.27871184494595,-0.90435610879647,-0.904906357718851,-0.999297694813687,0.322332299521183,0.363362291297433],
'G':[0.480108668470425,0.745340966903249,0.062916335574921,-0.218174540436266,0.250882292977333,-0.553429077935438,-1.80091468934086,-2.03184082539393,-1.7465820600918,0.912075945025212,-0.631383025410886,-0.999297694813687,-0.30514124354672,-0.837698138429041],
'H':[-0.400090557058688,0.17686056841772,-0.188749006724763,-0.225420646748007,0.771230752485876,-0.370490165573597,0.555901120322853,0.447282365999688,0.322666584912373,-0.304933531035315,1.0097569684369,1.00802518559809,0.262163603610562,0.440850060957206],
'I':[1.38031242185247,-1.41804277177779,-0.843078896703942,-0.224162112493862,-1.16149209711728,-0.545655469761422,0.105032008908926,-0.0186367652499869,0.702465386843518,-1.24493711888804,-1.26960413412947,-0.868385333047702,-1.14750298629541,0.700987573386442],
'K':[-1.50033958897008,0.745340966903249,1.57290838937303,-0.224314662100425,1.10574047645565,2.01186161948981,0.443183842469371,0.952208680330731,0.872720022191963,1.37526611874974,1.60239085510416,2.27351134933594,-0.743513170895529,-0.527747059789951],
'L':[1.06023997620552,-1.60753623793963,-0.843078896703942,-0.22301799044464,-1.27299533844054,-0.53010825341339,0.105032008908926,0.243578187685877,0.702465386843518,-0.972472310814783,-0.996080801821506,-0.737472971281717,-0.700535530959371,1.24340196100485],
'M':[0.6401448912939,-0.817980128931955,-0.591413554404258,-0.224886723125036,-0.975653361578516,-0.47932068000982,0.463677892988186,0.466785957540372,0.718181199337221,-0.768123704759844,-0.631383025410886,-0.562923155593736,-1.10452534635925,-1.10337049154826],
'N':[-0.780176586264441,0.950625555245245,1.06957770477366,-0.224810448321755,1.21724371777891,-0.378263773747613,-0.42781330458026,-0.354531952895101,-0.209051737791231,0.871206223814225,0.872995302282922,0.702563008144121,2.74627119192048,-1.38011252604745],
'P':[0.120027167117606,0.17686056841772,0.062916335574921,-0.215848158936181,-0.120795178100197,-0.553429077935438,-0.458554380358482,-0.759773243795982,-0.649094487614904,0.303571206994949,-0.585795803359558,-0.781110425203712,-0.520029443227509,0.213921592667873],
'Q':[-0.850192433749711,0.792714333443709,0.163582472494795,-0.223132402649562,0.808398499593628,-0.370490165573597,0.0435498573524813,0.245745253412619,0.367194720311197,0.603282495875526,1.0097569684369,1.00802518559809,0.262163603610562,-0.0572856011413323],
'R':[-2.5305727733962,0.508474134200945,1.57290838937303,-0.223323089657766,0.808398499593628,2.1414217557234,1.18096966114671,1.60666252980702,1.00368512630615,1.33439639753875,1.51121641100151,1.48803717874003,-0.820872922780613,-0.843232979119025],
'S':[-0.180040750676409,0.871669944344477,0.213915540954732,-0.224810448321755,0.325217787192839,-0.466882906931394,-1.16559912325759,-1.12817441734224,-0.979126549982658,1.42975908036439,0.143599749461682,-0.0392737085297955,1.31941354604004,-0.073890123211283],
'T':[-0.050011319632336,0.666385356002481,-0.138415938264826,-0.224886723125036,0.102211304546321,-0.467401147476329,-0.694235961324853,-0.636250497371649,-0.40288009188023,0.603282495875526,0.371535859718319,-0.0392737085297955,0.399692051406267,-0.64397871427961],
'V':[1.08024450405846,-0.739024518031187,-0.692079691324131,-0.222827303436436,-0.90131786736301,-0.546691950851291,-0.366331153023816,-0.376202610162528,0.12621892874109,-0.727253983548856,-1.17842969002682,-0.824747879125707,-0.545816027189203,1.97953577277269],
'W':[0.810183378043843,-1.73386521538086,-1.64840799206293,-0.22355191406761,-1.08715660290178,-0.444598563499216,2.39011864175678,1.82987029966151,2.06450246963108,-1.64909325086336,-0.494621359256903,-0.693835517359722,-0.666153419010445,-0.73253616531935],
'Y':[0.260058862088147,-0.454784318788423,-1.09474423900363,4.2485160771289,-0.789814626039751,-0.469992350201001,1.25269883796256,1.19058591027243,-1.74595342759205,-1.43112140440476,0.645059192026285,0.35346337676816,-0.0816575158786997,0.750801139596297],
}
def ext_prop_enc(sequence):
temp_list = []
for letter in sequence:
temp_list.append(letter)
new_list = []
for i,letter in enumerate(temp_list):
new_list.append(extended_prop_aa[temp_list[i]])
return np.array(new_list).reshape(-1)
seq_rep['PhysProperties'] = [ext_prop_enc(seq) for seq in seq_list]
seq_rep['PhysProperties'][0]
array([ 1.0802445 , -0.73902452, -0.69207969, -0.2228273 , -0.90131787,
-0.54669195, -0.36633115, -0.37620261, 0.12621893, -0.72725398,
-1.17842969, -0.82474788, -0.54581603, 1.97953577, 0.81018338,
-1.73386522, -1.64840799, -0.22355191, -1.0871566 , -0.44459856,
2.39011864, 1.8298703 , 2.06450247, -1.64909325, -0.49462136,
-0.69383552, -0.66615342, -0.73253617, -2.53057277, 0.50847413,
1.57290839, -0.22332309, 0.8083985 , 2.14142176, 1.18096966,
1.60666253, 1.00368513, 1.3343964 , 1.51121641, 1.48803718,
-0.82087292, -0.84323298, -0.90020375, 1.42435922, 1.57290839,
-0.22591643, 1.73759218, 1.55580994, -0.72497704, -0.50189242,
-0.69886123, 0.71226842, 0.73623364, 0.74620046, 1.58587491,
-0.88197686, 0.62014036, 0.17686057, -0.18874901, -0.22473417,
-0.08362743, -0.55342908, -1.32955153, -1.38172111, -1.02627399,
-0.50928214, -0.5857958 , -0.78111043, -0.52002944, 0.21392159,
-0.74016753, 1.07695453, 1.57290839, -0.22473417, 1.47741795,
2.03310948, -0.25361388, 0.09405065, -0.12261477, 0.79400786,
1.00975697, 0.96438773, -0.62317578, -0.96499947, -0.90020375,
1.42435922, 1.57290839, -0.22591643, 1.73759218, 1.55580994,
-0.72497704, -0.50189242, -0.69886123, 0.71226842, 0.73623364,
0.74620046, 1.58587491, -0.88197686, 0.26005886, -0.45478432,
-1.09474424, 4.24851608, -0.78981463, -0.46999235, 1.25269884,
1.19058591, -1.74595343, -1.4311214 , 0.64505919, 0.35346338,
-0.08165752, 0.75080114, 0.62014036, 0.17686057, -0.18874901,
-0.22473417, -0.08362743, -0.55342908, -1.32955153, -1.38172111,
-1.02627399, -0.50928214, -0.5857958 , -0.78111043, -0.52002944,
0.21392159, -0.05001132, 0.66638536, -0.13841594, -0.22488672,
0.1022113 , -0.46740115, -0.69423596, -0.6362505 , -0.40288009,
0.6032825 , 0.37153586, -0.03927371, 0.39969205, -0.64397871,
1.19026941, -1.60753624, -1.19541038, -0.22355191, -1.1614921 ,
-0.53529066, 1.17072264, 0.88719671, 1.27871184, -0.90435611,
-0.90490636, -0.99929769, 0.3223323 , 0.36336229, 0.12002717,
0.17686057, 0.06291634, -0.21584816, -0.12079518, -0.55342908,
-0.45855438, -0.75977324, -0.64909449, 0.30357121, -0.5857958 ,
-0.78111043, -0.52002944, 0.21392159])
NGRAM representation
N-grams encoding was completed by pre-calculating the observed n-mers in the dataset up to a maximum n-mer length of the full peptide length (12 residues). The entire peptide was represented at once as a 138,622 length vector, where each index of the vector describes an n-mer motif that is either present (1) or absent (0) in the peptide.
# minlengthfinder will be used to identify the minimum peptide length from the input dataset
# which is 12
def minlengthfinder(lst):
lengths = []
for x in lst: lengths.append(len(x))
the_min = min(lengths)
return the_min
NGRAM_MAX = minlengthfinder(seq_list)
print(f'NGRAM_MAX is {NGRAM_MAX}, now calculating n_mers (the index of the N-grams vector)',flush=True)
NGRAM_MAX is 12, now calculating n_mers (the index of the N-grams vector)
def cal_n_mers(seq_list, n): # enumerate and index of all the unique n_mers in the input dataset
theo_max = len(Std_All_AA_Smiles)**n
n_mers = set()
for seq in seq_list:
all_mers = [seq[i:i+n] for i in range(len(seq)-n+1)]
n_mers.update(all_mers)
if n_mers == theo_max:
print('NGrams pre-calculation reached the theoretical maximum and stopped')
break
return n_mers
# n-mers are calculated here, if additional peptides are added, the same n-mers should be used
n_mers = [list(cal_n_mers(seq_list, n)) for n in range(1, NGRAM_MAX+1)]
def n_grams(seq,n_mers):
n_gram = []
for n_mer in n_mers:
n = len(n_mer[0])
seq_mers = [seq[i:i+n] for i in range(len(seq)-n+1)]
for mer in n_mer:
n_gram.append(seq_mers.count(mer))
return np.array(n_gram)
def enc_ngrams(seqs,n_mers):
return [n_grams(seq,n_mers) for seq in seqs]
seq_rep['NGrams'] = enc_ngrams(seq_list,n_mers)
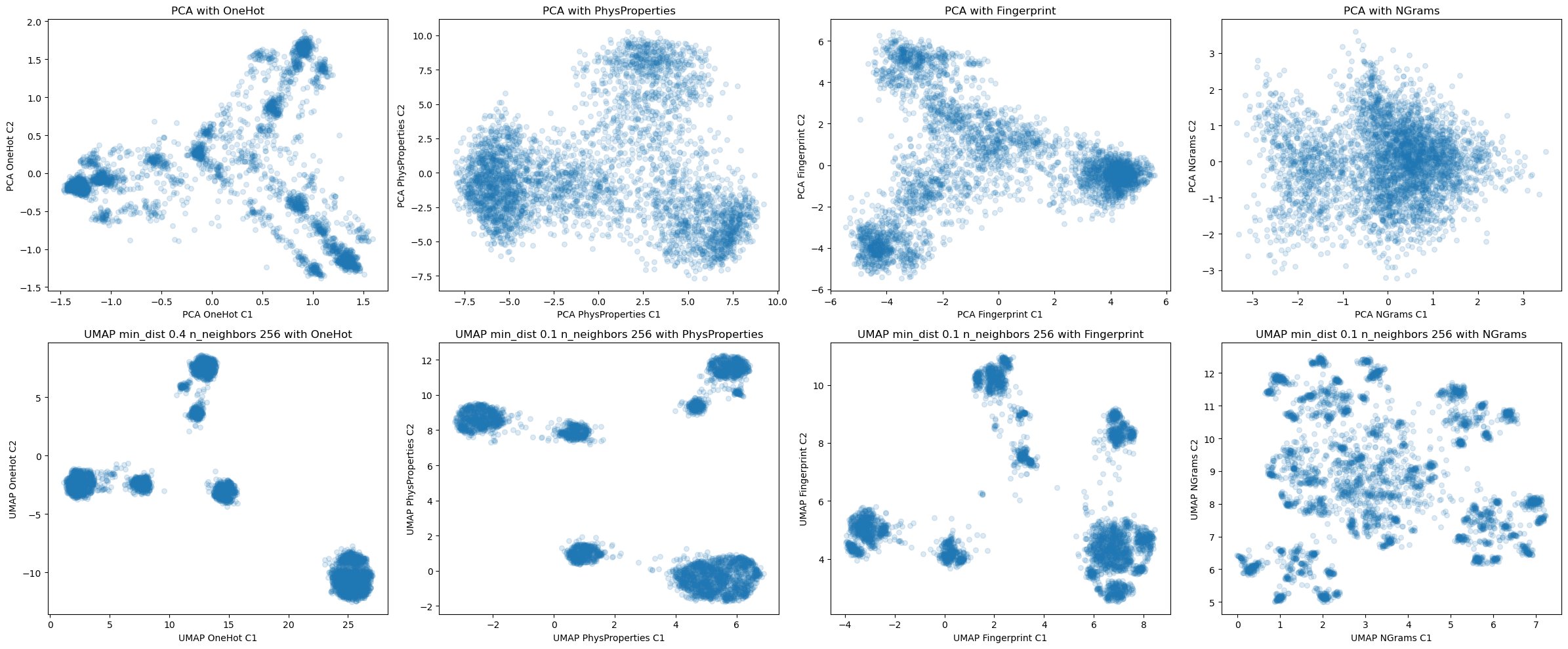
Dim Reduction with PCA
import matplotlib.pyplot as plt
from sklearn import decomposition
data = {}
SEED = 108
pca_fn_dict = {}
pca_embeddding_dict = {}
for rep in seq_rep:
ref_feature = seq_rep[rep]
reducer_scaled_dict = {}
embedding_scaled_dict = {}
pca = decomposition.PCA(n_components=2, random_state=SEED)
embedding = pca.fit_transform(ref_feature)
pca_fn_dict[rep] = pca
pca_embeddding_dict[rep] = embedding
data['PCA'] = pca_embeddding_dict
Dim Reduction with UMAP
#!pip install umap-learn
from sklearn import manifold
import umap.umap_ as umap
# Dictionaries will hold the data, format of umap_all[min_dist][representation][n_neighbors]
umap_all = {}
umap_fn_dict = {}
umap_embeddding_dict = {}
n_bors = [8] #to determine if UMAP embeddings are stable
min_dict = {'OneHot': 0.4, 'Fingerprint': 0.1, 'PhysProperties': 0.1, 'NGrams': 0.1}
metric_dict = {'OneHot': 'rogerstanimoto', 'Fingerprint': 'rogerstanimoto', 'PhysProperties': 'euclidean', 'NGrams': 'rogerstanimoto'}
for rep in seq_rep: # iterate over representation
min_num = min_dict[rep]
print(f'min_num is {min_num} for {rep}')
ref_feature = seq_rep[rep]
reducer_scaled_dict = {}
embedding_scaled_dict = {}
for n_neighbors in n_bors: # iterate over n_neighbors if n_bors is constructed with multiple numbers
print(f'n_neighbors is {n_neighbors}',flush=True)
reducer_scaled_dict[2**n_neighbors] = umap.UMAP(
n_components=2, n_neighbors=2**n_neighbors, min_dist=min_dict[rep], random_state=SEED, metric=metric_dict[rep])
embedding_scaled_dict[2**n_neighbors] = reducer_scaled_dict[2**n_neighbors].fit_transform(ref_feature)
umap_fn_dict[rep] = reducer_scaled_dict
umap_embeddding_dict[rep] = embedding_scaled_dict
umap_all[min_num] = umap_embeddding_dict
data['UMAP'] = umap_all
min_num is 0.4 for OneHot
n_neighbors is 8
/Users/sasha/anaconda3/lib/python3.10/site-packages/umap/umap_.py:1879: UserWarning: gradient function is not yet implemented for rogerstanimoto distance metric; inverse_transform will be unavailable
warn(
/Users/sasha/anaconda3/lib/python3.10/site-packages/umap/umap_.py:1943: UserWarning: n_jobs value -1 overridden to 1 by setting random_state. Use no seed for parallelism.
warn(f"n_jobs value {self.n_jobs} overridden to 1 by setting random_state. Use no seed for parallelism.")
min_num is 0.1 for Fingerprint
n_neighbors is 8
/Users/sasha/anaconda3/lib/python3.10/site-packages/umap/umap_.py:1879: UserWarning: gradient function is not yet implemented for rogerstanimoto distance metric; inverse_transform will be unavailable
warn(
/Users/sasha/anaconda3/lib/python3.10/site-packages/umap/umap_.py:1943: UserWarning: n_jobs value -1 overridden to 1 by setting random_state. Use no seed for parallelism.
warn(f"n_jobs value {self.n_jobs} overridden to 1 by setting random_state. Use no seed for parallelism.")
min_num is 0.1 for PhysProperties
n_neighbors is 8
/Users/sasha/anaconda3/lib/python3.10/site-packages/umap/umap_.py:1943: UserWarning: n_jobs value -1 overridden to 1 by setting random_state. Use no seed for parallelism.
warn(f"n_jobs value {self.n_jobs} overridden to 1 by setting random_state. Use no seed for parallelism.")
min_num is 0.1 for NGrams
n_neighbors is 8
/Users/sasha/anaconda3/lib/python3.10/site-packages/umap/umap_.py:1879: UserWarning: gradient function is not yet implemented for rogerstanimoto distance metric; inverse_transform will be unavailable
warn(
/Users/sasha/anaconda3/lib/python3.10/site-packages/umap/umap_.py:1943: UserWarning: n_jobs value -1 overridden to 1 by setting random_state. Use no seed for parallelism.
warn(f"n_jobs value {self.n_jobs} overridden to 1 by setting random_state. Use no seed for parallelism.")
# Choose dimensionality techniques
Available_Dim_red = {'PCA': True, 'UMAP': True}
dim_red_list = []
for key,value in Available_Dim_red.items():
if bool(value): dim_red_list.append(key)
print(dim_red_list)
['PCA', 'UMAP']
def lister(list1,list2): # Makes combinations of each element in both lists for plotting
out = []
for i in list1:
for l in list2:
out.append((i,l))
return out
fig, axes = plt.subplots(len(dim_red_list),len(seq_rep_list),figsize = (6*len(seq_rep_list),5*len(dim_red_list)))
reps_plotting = lister(dim_red_list,seq_rep_list)
for ax, (red,enc) in zip(fig.axes,reps_plotting):
print(enc,red)
if red == 'UMAP':
C1 = data[red][min_dict[enc]][enc][2**n_bors[0]][:,0]
C2 = data[red][min_dict[enc]][enc][2**n_bors[0]][:,1]
ax.set_title(f'{red} min_dist {min_dict[enc]} n_neighbors {2**n_bors[0]} with {enc}')
else:
C1 = data[red][enc][:,0]
C2 = data[red][enc][:,1]
ax.set_title(f'{red} with {enc}')
plot = ax.scatter(C1,C2,s=30,marker='o',alpha=0.15,c='tab:blue')
ax.set_xlabel(f'{red} {enc} C1')
ax.set_ylabel(f'{red} {enc} C2')
plt.tight_layout()
#plt.savefig(f'{Savetitle} DimRed Embeddings.png',dpi=300)
plt.show
OneHot PCA
PhysProperties PCA
Fingerprint PCA
NGrams PCA
OneHot UMAP
PhysProperties UMAP
Fingerprint UMAP
NGrams UMAP
<function matplotlib.pyplot.show(close=None, block=None)>
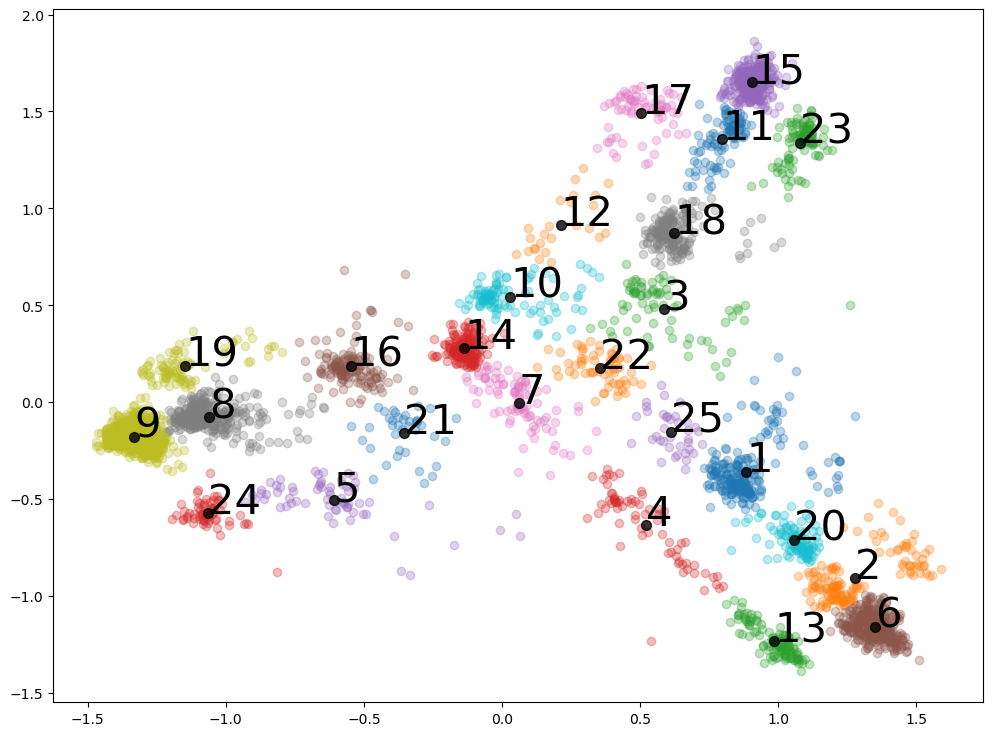
Looking at clusters - one from PCA one hot encoding
from sklearn.cluster import AgglomerativeClustering
from matplotlib import pyplot as plt
df1 = pd.read_csv('./PCA OneHot.csv')
df1.head()
| Peptide | Label | PCA OneHot C1 | PCA OneHot C2 | |
|---|---|---|---|---|
| 0 | LPWKWPPFYERF | Nonspecific | 0.668123 | -0.103624 |
| 1 | FFFYQMYPWWAY | Nonspecific | -0.020174 | 0.059053 |
| 2 | FSVAPHWLWTYH | Nonspecific | 0.413959 | 0.273859 |
| 3 | WLYLPYWTEHPQ | Nonspecific | 0.386628 | 0.191751 |
| 4 | WGQMFTWFDHQP | Nonspecific | -0.046315 | 0.104411 |
all_column_list = df1.columns.to_list()
C1_col_name = [s for s in all_column_list if 'C1' in s][0]
C2_col_name = [s for s in all_column_list if 'C2' in s][0]
X = np.asarray([(float(x), float(y)) for x, y in zip(df1[C1_col_name], df1[C2_col_name])])
X.shape
(4104, 2)
eps_range = ['']
min_samp_range_start = [25]
for count1,epsy in enumerate(eps_range):
for count2,min_samp in enumerate(min_samp_range_start):
model = AgglomerativeClustering(n_clusters=min_samp) # define the model
print(f'AggCl detection, n_clusters is {str(min_samp)}')
epsy = ''
yhat = model.fit_predict(X) # fit model, predict clusters. Every row has a cluster associated.
yhat = yhat + 1 # This is done because clusters start at 0, and we want to label them starting at 1.
clusters = np.unique(yhat)
Center_ave = np.zeros([len(clusters)+1,2]) # +1 is to make both DBSCAN (which has a noise cluster) and AggCl (which does not have a noise cluster) comptabile
fig,ax = plt.subplots()
fig.set_size_inches(12, 9)
for j,i in enumerate(clusters):
row_ix = np.where(yhat == i) # get row indexes for samples with this cluster
if i != 0:
# Plot the data
plt.scatter(X[row_ix, 0], X[row_ix, 1], alpha = 0.3)
# Plot points that are at the center of each cluster, and annotate them with their autonomous number label
Center_ave[i,0] = np.average(X[row_ix, 0])
Center_ave[i,1] = np.average(X[row_ix, 1])
plt.scatter(Center_ave[i,0], Center_ave[i,1], alpha=0.8, s=50, color='black')
plt.annotate(i, (Center_ave[i,0], Center_ave[i,1]), fontsize = 30, alpha=1)
if i == 0:
plt.scatter(X[row_ix, 0], X[row_ix, 1], alpha = 0.3, color='gray') # plot the noise points less intensely
AggCl detection, n_clusters is 25