Pharmocophore Matching
Posted on * • 5 minutes • 1005 words
A pharmacophore is a hypothetical description of the molecular features necessary for a ligand to bind to a receptor and exert a biological response. In simpler terms, it’s the “template” of a molecule that helps it interact with a target in a specific way. These features can include hydrogen bond donors and acceptors, aromatic rings, positively or negatively charged groups, and hydrophobic regions.
Pharmacophore models are used in drug design and discovery to identify and optimize molecules that can bind to a target protein or receptor with high affinity and specificity. They are essential tools in computer-aided drug design (CADD) where computational methods are used to screen and design potential drug candidates and understand the structure-activity relationships (SAR) of molecules and guide the development of new therapeutic agents.
import pandas as pd
# Load SMILES for PDB ligand structures
ligands = pd.read_csv("./PDB_top_ligands.csv")
ligands
| @structureId | @chemicalID | @type | @molecularWeight | chemicalName | formula | InChI | InChIKey | smiles | |
|---|---|---|---|---|---|---|---|---|---|
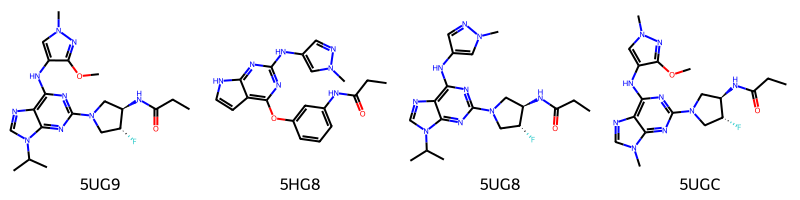
| 0 | 5UG9 | 8AM | NON-POLYMER | 445.494 | N-[(3R,4R)-4-fluoro-1-{6-[(3-methoxy-1-methyl-... | C20 H28 F N9 O2 | InChI=1S/C20H28FN9O2/c1-6-15(31)23-13-9-29(7-1... | MJLFLAORJNTNDV-CHWSQXEVSA-N | CCC(=O)N[C@@H]1CN(C[C@H]1F)c2nc(c3c(n2)n(cn3)C... |
| 1 | 5HG8 | 634 | NON-POLYMER | 377.400 | N-[3-({2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-... | C19 H19 N7 O2 | InChI=1S/C19H19N7O2/c1-3-16(27)22-12-5-4-6-14(... | YWNHZBNRKJYHTR-UHFFFAOYSA-N | CCC(=O)Nc1cccc(c1)Oc2c3cc[nH]c3nc(n2)Nc4cnn(c4)C |
| 2 | 5UG8 | 8BP | NON-POLYMER | 415.468 | N-[(3R,4R)-4-fluoro-1-{6-[(1-methyl-1H-pyrazol... | C19 H26 F N9 O | InChI=1S/C19H26FN9O/c1-5-15(30)24-14-9-28(8-13... | CGULPICMFDDQRH-ZIAGYGMSSA-N | CCC(=O)N[C@@H]1CN(C[C@H]1F)c2nc(c3c(n2)n(cn3)C... |
| 3 | 5UGC | 8BS | NON-POLYMER | 417.441 | N-[(3R,4R)-4-fluoro-1-{6-[(3-methoxy-1-methyl-... | C18 H24 F N9 O2 | InChI=1S/C18H24FN9O2/c1-5-13(29)21-11-8-28(6-1... | XWNKXCUQRQRAFF-GHMZBOCLSA-N | CCC(=O)N[C@@H]1CN(C[C@H]1F)c2nc(c3c(n2)n(cn3)C... |
from rdkit import RDConfig, Chem, Geometry, DistanceGeometry
from rdkit.Chem import ChemicalFeatures, rdDistGeom, Draw, rdMolTransforms, AllChem
from rdkit.Chem.Draw import DrawingOptions
from rdkit.Chem.Pharm3D import Pharmacophore, EmbedLib
from rdkit.Numerics import rdAlignment
smils = [Chem.MolFromSmiles(x) for x in ligands['smiles'].tolist()]
Draw.MolsToGridImage(
smils,
molsPerRow=4,
legends=ligands['@structureId'].tolist()
)
Generate Conformers
ms = [Chem.AddHs(m) for m in smils]
ps = AllChem.ETKDGv3()
ps.randomSeed = 0xf00d # we seed the RNG so that this is reproducible
for m in ms:
AllChem.EmbedMolecule(m,ps)
Features of pharmacophore
import os.path
fdef = os.path.join(RDConfig.RDDataDir,'BaseFeatures.fdef')
featFactory = AllChem.BuildFeatureFactory(fdef)
#There are many features
list(featFactory.GetFeatureDefs().keys())
['Donor.SingleAtomDonor',
'Acceptor.SingleAtomAcceptor',
'NegIonizable.AcidicGroup',
'PosIonizable.BasicGroup',
'PosIonizable.PosN',
'PosIonizable.Imidazole',
'PosIonizable.Guanidine',
'ZnBinder.ZnBinder1',
'ZnBinder.ZnBinder2',
'ZnBinder.ZnBinder3',
'ZnBinder.ZnBinder4',
'ZnBinder.ZnBinder5',
'ZnBinder.ZnBinder6',
'Aromatic.Arom4',
'Aromatic.Arom5',
'Aromatic.Arom6',
'Aromatic.Arom7',
'Aromatic.Arom8',
'Hydrophobe.ThreeWayAttach',
'Hydrophobe.ChainTwoWayAttach',
'LumpedHydrophobe.Nitro2',
'LumpedHydrophobe.RH6_6',
'LumpedHydrophobe.RH5_5',
'LumpedHydrophobe.RH4_4',
'LumpedHydrophobe.RH3_3',
'LumpedHydrophobe.tButyl',
'LumpedHydrophobe.iPropyl']
print(featFactory.GetFeatureFamilies())
('Donor', 'Acceptor', 'NegIonizable', 'PosIonizable', 'ZnBinder', 'Aromatic', 'Hydrophobe', 'LumpedHydrophobe')
ligand_smiles = [ligands["smiles"].values]
ligand_smiles
[array(['CCC(=O)N[C@@H]1CN(C[C@H]1F)c2nc(c3c(n2)n(cn3)C(C)C)Nc4cn(nc4OC)C',
'CCC(=O)Nc1cccc(c1)Oc2c3cc[nH]c3nc(n2)Nc4cnn(c4)C',
'CCC(=O)N[C@@H]1CN(C[C@H]1F)c2nc(c3c(n2)n(cn3)C(C)C)Nc4cnn(c4)C',
'CCC(=O)N[C@@H]1CN(C[C@H]1F)c2nc(c3c(n2)n(cn3)C)Nc4cn(nc4OC)C'],
dtype=object)]
print(Chem.MolToSmiles(ms[0]))
[H]c1c(N([H])c2nc(N3C([H])([H])[C@@]([H])(F)[C@]([H])(N([H])C(=O)C([H])([H])C([H])([H])[H])C3([H])[H])nc3c2nc([H])n3C([H])(C([H])([H])[H])C([H])([H])[H])c(OC([H])([H])[H])nn1C([H])([H])[H]
# NBVAL_CHECK_OUTPUT
molecules = []
for mol in ms:
Chem.SanitizeMol(mol)
print(Chem.MolToSmiles(mol))
molecules.append(mol)
print(f"Number of molecules: {len(molecules)}")
[H]c1c(N([H])c2nc(N3C([H])([H])[C@@]([H])(F)[C@]([H])(N([H])C(=O)C([H])([H])C([H])([H])[H])C3([H])[H])nc3c2nc([H])n3C([H])(C([H])([H])[H])C([H])([H])[H])c(OC([H])([H])[H])nn1C([H])([H])[H]
[H]c1nn(C([H])([H])[H])c([H])c1N([H])c1nc(Oc2c([H])c([H])c([H])c(N([H])C(=O)C([H])([H])C([H])([H])[H])c2[H])c2c([H])c([H])n([H])c2n1
[H]c1nn(C([H])([H])[H])c([H])c1N([H])c1nc(N2C([H])([H])[C@@]([H])(F)[C@]([H])(N([H])C(=O)C([H])([H])C([H])([H])[H])C2([H])[H])nc2c1nc([H])n2C([H])(C([H])([H])[H])C([H])([H])[H]
[H]c1c(N([H])c2nc(N3C([H])([H])[C@@]([H])(F)[C@]([H])(N([H])C(=O)C([H])([H])C([H])([H])[H])C3([H])[H])nc3c2nc([H])n3C([H])([H])[H])c(OC([H])([H])[H])nn1C([H])([H])[H]
Number of molecules: 4
feature_colors = {
"donors": (0, 0.9, 0), # Green
"acceptors": (0.9, 0, 0), # Red
"hydrophobics": (1, 0.9, 0), # Yellow
}
import nglview as nv
import time
def show_ligands(molecules):
"""Generate a view of the ligand molecules.
Parameters
-----------
molecules: list of rdkit.Chem.rdchem.Mol
Returns
----------
nglview.widget.NGLWidget
"""
view = nv.NGLWidget()
for molecule in molecules:
component = view.add_component(molecule)
time.sleep(0.1)
component.clear()
component.add_ball_and_stick(multipleBond=True)
return view
def visualize_features(
molecules,
features,
feature_type="features",
color="yellow",
sphere_radius=0.5,
):
"""Generate a view of the molecules highlighting the specified feature type.
Parameters
-----------
molecules: list of rdkit.Chem.rdchem.Mol
molecules to be visualized
features: list of tuples of rdkit.Chem.rdMolChemicalFeatures.MolChemicalFeature
extracted features from molecules chosen to be highlighted
feature_type: string, optional
name of the feature to be highlighted
color: string, optional
color used to display the highlighted features
sphere_radius: float, optional
display size of the highlighted features
Returns
----------
nglview.widget.NGLWidget
"""
print(f"Number of {feature_type} in all ligands: {sum([len(i) for i in features])}")
view = show_ligands(molecules)
for i, feature_set in enumerate(features, 1):
for feature in feature_set:
loc = list(feature.GetPos())
label = f"{feature_type}_{i}"
view.shape.add_sphere(loc, color, sphere_radius, label)
return view
features = featFactory.GetFeaturesForMol(ms[0])
print(f"Number of features found: {len(features)}")
Number of features found: 18
acceptors = []
donors = []
hydrophobics = []
for molecule in molecules:
acceptors.append(featFactory.GetFeaturesForMol(molecule, includeOnly="Acceptor"))
donors.append(featFactory.GetFeaturesForMol(molecule, includeOnly="Donor"))
hydrophobics.append(featFactory.GetFeaturesForMol(molecule, includeOnly="Hydrophobe"))
featDic = {
"donors": donors,
"acceptors": acceptors,
"hydrophobics": hydrophobics,
}
feature_type = "donors"
view = visualize_features(
molecules,
featDic[feature_type],
feature_type,
feature_colors[feature_type],
)
view
Number of donors in all ligands: 10
NGLWidget()
features[1]
<rdkit.Chem.rdMolChemicalFeatures.MolChemicalFeature at 0x12f78aea0>
import collections
from collections import Counter
feature_frequency = collections.Counter(sorted([feature.GetFamily() for feature in features]))
feature_frequency
Counter({'Acceptor': 7,
'Aromatic': 3,
'Donor': 2,
'Hydrophobe': 4,
'LumpedHydrophobe': 1,
'PosIonizable': 1})
Visualize features
features = featFactory.GetFeaturesForMol(ms[0])
pcophore = Pharmacophore.Pharmacophore(features)
Compare Pharmocophore with a different ligand smiles
ligand = Chem.MolFromSmiles("c1ccc(-c2n[nH]cc2-c2ccnc3ccccc23)nc1")
DrawingOptions.bondLineWidth=1.8
DrawingOptions.atomLabelFontSize=14
DrawingOptions.includeAtomNumbers=True
ligand
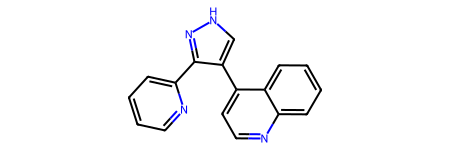
canMatch,allMatches = EmbedLib.MatchPharmacophoreToMol(ligand,featFactory,pcophore)
canMatch
False
Compare Pharmocophore with a similar ligand smiles
ligand = smils[2]
ligand
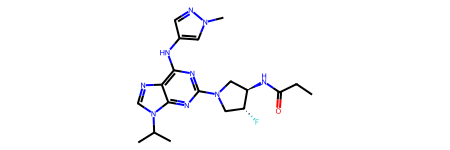
canMatch,allMatches = EmbedLib.MatchPharmacophoreToMol(ligand,featFactory,pcophore)
canMatch
True
for (i,match) in enumerate(allMatches):
for f in match:
print("%d %s %s %s"%(i, f.GetFamily(), f.GetType(), f.GetAtomIds()))
0 Donor SingleAtomDonor (4,)
0 Donor SingleAtomDonor (23,)
1 Donor SingleAtomDonor (4,)
1 Donor SingleAtomDonor (23,)
2 Acceptor SingleAtomAcceptor (3,)
2 Acceptor SingleAtomAcceptor (10,)
2 Acceptor SingleAtomAcceptor (12,)
2 Acceptor SingleAtomAcceptor (16,)
2 Acceptor SingleAtomAcceptor (19,)
2 Acceptor SingleAtomAcceptor (26,)
3 Acceptor SingleAtomAcceptor (3,)
3 Acceptor SingleAtomAcceptor (10,)
3 Acceptor SingleAtomAcceptor (12,)
3 Acceptor SingleAtomAcceptor (16,)
3 Acceptor SingleAtomAcceptor (19,)
3 Acceptor SingleAtomAcceptor (26,)
4 Acceptor SingleAtomAcceptor (3,)
4 Acceptor SingleAtomAcceptor (10,)
4 Acceptor SingleAtomAcceptor (12,)
4 Acceptor SingleAtomAcceptor (16,)
4 Acceptor SingleAtomAcceptor (19,)
4 Acceptor SingleAtomAcceptor (26,)
5 Acceptor SingleAtomAcceptor (3,)
5 Acceptor SingleAtomAcceptor (10,)
5 Acceptor SingleAtomAcceptor (12,)
5 Acceptor SingleAtomAcceptor (16,)
5 Acceptor SingleAtomAcceptor (19,)
5 Acceptor SingleAtomAcceptor (26,)
6 Acceptor SingleAtomAcceptor (3,)
6 Acceptor SingleAtomAcceptor (10,)
6 Acceptor SingleAtomAcceptor (12,)
6 Acceptor SingleAtomAcceptor (16,)
6 Acceptor SingleAtomAcceptor (19,)
6 Acceptor SingleAtomAcceptor (26,)
7 Acceptor SingleAtomAcceptor (3,)
7 Acceptor SingleAtomAcceptor (10,)
7 Acceptor SingleAtomAcceptor (12,)
7 Acceptor SingleAtomAcceptor (16,)
7 Acceptor SingleAtomAcceptor (19,)
7 Acceptor SingleAtomAcceptor (26,)
8 Acceptor SingleAtomAcceptor (3,)
8 Acceptor SingleAtomAcceptor (10,)
8 Acceptor SingleAtomAcceptor (12,)
8 Acceptor SingleAtomAcceptor (16,)
8 Acceptor SingleAtomAcceptor (19,)
8 Acceptor SingleAtomAcceptor (26,)
9 PosIonizable Imidazole (14, 19, 18, 17, 15)
10 Aromatic Arom5 (14, 15, 17, 18, 19)
10 Aromatic Arom5 (24, 25, 26, 27, 28)
10 Aromatic Arom6 (11, 12, 13, 14, 15, 16)
11 Aromatic Arom5 (14, 15, 17, 18, 19)
11 Aromatic Arom5 (24, 25, 26, 27, 28)
11 Aromatic Arom6 (11, 12, 13, 14, 15, 16)
12 Aromatic Arom5 (14, 15, 17, 18, 19)
12 Aromatic Arom5 (24, 25, 26, 27, 28)
12 Aromatic Arom6 (11, 12, 13, 14, 15, 16)
13 Hydrophobe ChainTwoWayAttach (1,)
14 Hydrophobe ChainTwoWayAttach (1,)
15 Hydrophobe ChainTwoWayAttach (1,)
16 Hydrophobe ChainTwoWayAttach (1,)
17 LumpedHydrophobe iPropyl (20, 21, 22)
Reference:
Talktutorial - T009 by Volakmer Lab
https://greglandrum.github.io/rdkit-blog/posts/2023-02-24-using-feature-maps.html
https://github.com/rdkit/UGM_2016/blob/master/Notebooks/Stiefl_RDKitPh4FullPublication.ipynb