Solubility Prediction using GNN
Posted on * • 7 minutes • 1399 words
We will use ESOL dataset and train GNN model to predict solubility directly from chemical structures
! pip install rdkit-pypi
!python -c "import torch; print(torch.__version__)"
2.1.0+cu121
!pip install torch-scatter -f https://data.pyg.org/whl/torch-2.1.0+cu121.html
!pip install torch-sparse -f https://data.pyg.org/whl/torch-2.1.0+cu121.html
!pip install torch-geometric
Dataset
import rdkit
from torch_geometric.datasets import MoleculeNet
# Load the ESOL dataset
data = MoleculeNet(root=".", name="ESOL")
data
Downloading https://deepchemdata.s3-us-west-1.amazonaws.com/datasets/delaney-processed.csv
Processing...
Done!
ESOL(1128)
# Investigating the dataset
print("Dataset type: ", type(data))
print("Dataset features: ", data.num_features)
print("Dataset target: ", data.num_classes)
print("Dataset length: ", data.len())
print("Sample nodes: ", data[0].num_nodes)
print("Sample edges: ", data[0].num_edges)
Dataset type: <class 'torch_geometric.datasets.molecule_net.MoleculeNet'>
Dataset features: 9
Dataset target: 734
Dataset length: 1128
Sample nodes: 32
Sample edges: 68
print("Dataset sample: ", data[0])
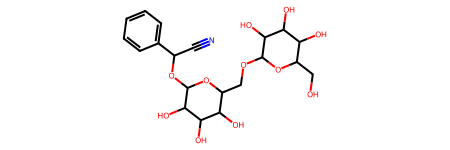
Dataset sample: Data(x=[32, 9], edge_index=[2, 68], edge_attr=[68, 3], smiles='OCC3OC(OCC2OC(OC(C#N)c1ccccc1)C(O)C(O)C2O)C(O)C(O)C3O ', y=[1, 1])
# Look at features
data[0].x
tensor([[8, 0, 2, 5, 1, 0, 4, 0, 0],
[6, 0, 4, 5, 2, 0, 4, 0, 0],
[6, 0, 4, 5, 1, 0, 4, 0, 1],
[8, 0, 2, 5, 0, 0, 4, 0, 1],
[6, 0, 4, 5, 1, 0, 4, 0, 1],
[8, 0, 2, 5, 0, 0, 4, 0, 0],
[6, 0, 4, 5, 2, 0, 4, 0, 0],
[6, 0, 4, 5, 1, 0, 4, 0, 1],
[8, 0, 2, 5, 0, 0, 4, 0, 1],
[6, 0, 4, 5, 1, 0, 4, 0, 1],
[8, 0, 2, 5, 0, 0, 4, 0, 0],
[6, 0, 4, 5, 1, 0, 4, 0, 0],
[6, 0, 2, 5, 0, 0, 2, 0, 0],
[7, 0, 1, 5, 0, 0, 2, 0, 0],
[6, 0, 3, 5, 0, 0, 3, 1, 1],
[6, 0, 3, 5, 1, 0, 3, 1, 1],
[6, 0, 3, 5, 1, 0, 3, 1, 1],
[6, 0, 3, 5, 1, 0, 3, 1, 1],
[6, 0, 3, 5, 1, 0, 3, 1, 1],
[6, 0, 3, 5, 1, 0, 3, 1, 1],
[6, 0, 4, 5, 1, 0, 4, 0, 1],
[8, 0, 2, 5, 1, 0, 4, 0, 0],
[6, 0, 4, 5, 1, 0, 4, 0, 1],
[8, 0, 2, 5, 1, 0, 4, 0, 0],
[6, 0, 4, 5, 1, 0, 4, 0, 1],
[8, 0, 2, 5, 1, 0, 4, 0, 0],
[6, 0, 4, 5, 1, 0, 4, 0, 1],
[8, 0, 2, 5, 1, 0, 4, 0, 0],
[6, 0, 4, 5, 1, 0, 4, 0, 1],
[8, 0, 2, 5, 1, 0, 4, 0, 0],
[6, 0, 4, 5, 1, 0, 4, 0, 1],
[8, 0, 2, 5, 1, 0, 4, 0, 0]])
# Investigating the edges in sparse COO format
data[0].edge_index.t()
tensor([[ 0, 1],
[ 1, 0],
[ 1, 2],
[ 2, 1],
[ 2, 3],
[ 2, 30],
[ 3, 2],
[ 3, 4],
[ 4, 3],
[ 4, 5],
[ 4, 26],
[ 5, 4],
[ 5, 6],
[ 6, 5],
[ 6, 7],
[ 7, 6],
[ 7, 8],
[ 7, 24],
[ 8, 7],
[ 8, 9],
[ 9, 8],
[ 9, 10],
[ 9, 20],
[10, 9],
[10, 11],
[11, 10],
[11, 12],
[11, 14],
[12, 11],
[12, 13],
[13, 12],
[14, 11],
[14, 15],
[14, 19],
[15, 14],
[15, 16],
[16, 15],
[16, 17],
[17, 16],
[17, 18],
[18, 17],
[18, 19],
[19, 14],
[19, 18],
[20, 9],
[20, 21],
[20, 22],
[21, 20],
[22, 20],
[22, 23],
[22, 24],
[23, 22],
[24, 7],
[24, 22],
[24, 25],
[25, 24],
[26, 4],
[26, 27],
[26, 28],
[27, 26],
[28, 26],
[28, 29],
[28, 30],
[29, 28],
[30, 2],
[30, 28],
[30, 31],
[31, 30]])
from rdkit import Chem
from rdkit.Chem.Draw import IPythonConsole
molecule = Chem.MolFromSmiles(data[0]["smiles"])
molecule
Graph Neural Network
import torch
from torch.nn import Linear
import torch.nn.functional as F
from torch_geometric.nn import GCNConv, TopKPooling, global_mean_pool
from torch_geometric.nn import global_mean_pool as gap, global_max_pool as gmp
embedding_size = 64
class GCN(torch.nn.Module):
def __init__(self):
# Init parent
super(GCN, self).__init__()
torch.manual_seed(42)
# GCN layers
self.initial_conv = GCNConv(data.num_features, embedding_size)
self.conv1 = GCNConv(embedding_size, embedding_size)
self.conv2 = GCNConv(embedding_size, embedding_size)
self.conv3 = GCNConv(embedding_size, embedding_size)
# Output layer
self.out = Linear(embedding_size*2, 1)
def forward(self, x, edge_index, batch_index):
# First Conv layer
hidden = self.initial_conv(x, edge_index)
hidden = F.tanh(hidden)
# Other Conv layers
hidden = self.conv1(hidden, edge_index)
hidden = F.tanh(hidden)
hidden = self.conv2(hidden, edge_index)
hidden = F.tanh(hidden)
hidden = self.conv3(hidden, edge_index)
hidden = F.tanh(hidden)
# Global Pooling (stack different aggregations)
hidden = torch.cat([gmp(hidden, batch_index),
gap(hidden, batch_index)], dim=1)
# Apply a final (linear) classifier.
out = self.out(hidden)
return out, hidden
model = GCN()
print(model)
GCN(
(initial_conv): GCNConv(9, 64)
(conv1): GCNConv(64, 64)
(conv2): GCNConv(64, 64)
(conv3): GCNConv(64, 64)
(out): Linear(in_features=128, out_features=1, bias=True)
)
Train
from torch_geometric.data import DataLoader
import warnings
warnings.filterwarnings("ignore")
# Root mean squared error
loss_fn = torch.nn.MSELoss()
optimizer = torch.optim.Adam(model.parameters(), lr=0.0007)
# Use GPU for training
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
model = model.to(device)
# Wrap data in a data loader
data_size = len(data)
NUM_GRAPHS_PER_BATCH = 64
loader = DataLoader(data[:int(data_size * 0.8)],
batch_size=NUM_GRAPHS_PER_BATCH, shuffle=True)
test_loader = DataLoader(data[int(data_size * 0.8):],
batch_size=NUM_GRAPHS_PER_BATCH, shuffle=True)
def train(data):
# Enumerate over the data
for batch in loader:
# Use GPU
batch.to(device)
# Reset gradients
optimizer.zero_grad()
# Passing the node features and the connection info
pred, embedding = model(batch.x.float(), batch.edge_index, batch.batch)
# Calculating the loss and gradients
loss = loss_fn(pred, batch.y)
loss.backward()
# Update using the gradients
optimizer.step()
return loss, embedding
print("Starting training...")
losses = []
for epoch in range(1000):
loss, h = train(data)
losses.append(loss)
if epoch % 100 == 0:
print(f"Epoch {epoch} | Train Loss {loss}")
Starting training...
Epoch 0 | Train Loss 0.7323977947235107
Epoch 100 | Train Loss 0.5643615126609802
Epoch 200 | Train Loss 0.8129488825798035
Epoch 300 | Train Loss 0.5515668988227844
Epoch 400 | Train Loss 0.26473188400268555
Epoch 500 | Train Loss 0.3548230826854706
Epoch 600 | Train Loss 0.10742906481027603
Epoch 700 | Train Loss 0.29880979657173157
Epoch 800 | Train Loss 0.08752292394638062
Epoch 900 | Train Loss 0.0839475765824318
Predictions
import pandas as pd
# Analyze the results for one batch
test_batch = next(iter(test_loader))
with torch.no_grad():
test_batch.to(device)
pred, embed = model(test_batch.x.float(), test_batch.edge_index, test_batch.batch)
df = pd.DataFrame()
df["y_real"] = test_batch.y.tolist()
df["y_pred"] = pred.tolist()
df["y_real"] = df["y_real"].apply(lambda row: row[0])
df["y_pred"] = df["y_pred"].apply(lambda row: row[0])
df
| y_real | y_pred | |
|---|---|---|
| 0 | -1.300 | -1.867198 |
| 1 | -3.953 | -4.592132 |
| 2 | -3.091 | -3.610150 |
| 3 | -2.210 | -2.107021 |
| 4 | -5.850 | -4.743058 |
| ... | ... | ... |
| 59 | -4.522 | -4.377628 |
| 60 | -4.286 | -1.482177 |
| 61 | -3.900 | -3.672484 |
| 62 | -5.060 | -4.966655 |
| 63 | -7.200 | -7.057222 |
64 rows × 2 columns
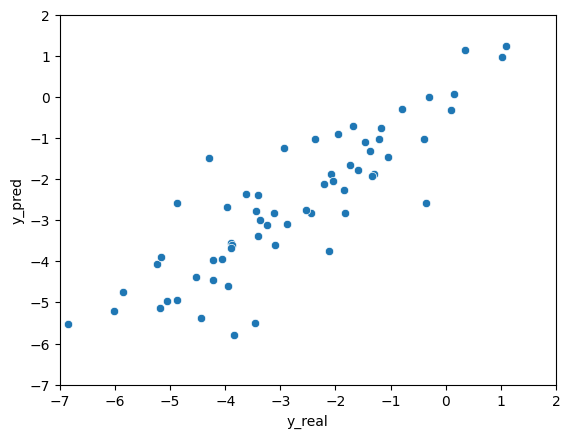
plt = sns.scatterplot(data=df, x="y_real", y="y_pred")
plt.set(xlim=(-7, 2))
plt.set(ylim=(-7, 2))
plt